Robust Multi-Layer Control Systems for Cooperative Cellular Behaviors
The goal of this project is to develop and demonstrate a multi-layer intra- and inter-cellular control systems integrated to create complex, spatially-organized, multi-functional model system for wound healing. Our system makes use of a layered control architecture with feedback at the DNA, RNA, protein, cellular and population levels to provide programmed phenotypic differentiation and interconnection between multiple cell types.
This project is an active collaboration with John Doyle, Michael Elowitz and Niles Pierce. This page describes the activities taking place in Richard Murray's group.
Project participants:
|
Collaborators:
|
Objectives
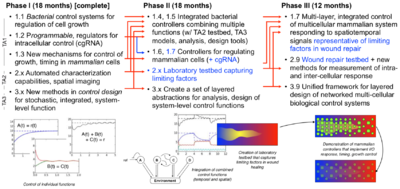
Phase I* objectives (Murray group):
- Biological controllers: Integrate multiple feedback controllers in E. coli, demonstrating ability to simultaneously modulate multiple input/output pairs.
- Testbeds: Expand micro- and macro-fluidic devicesfor temporal measurement and control of growth environments for integrated bacterial systems and individual mammalian systems.
- Testbeds: Conceptualize, design and develop a laboratory testbed for measurement and control of bacterial systems that is capable of emulating a wound-healing environment.
- Theory: Apply computational framework for sensor fusion to stochastic biological system data and integrate the results of the sensor fusion methods.
- Theory: Design an integrated feedback controller that uses time- scale separation to run a fast “inner” control loop and a slow “outer” control loop.
- Theory: Expand predictive models for multi-cellular systems in a cooperative control framework that allows robustness analysis and controller design of global input/output dynamics and interconnection structure.
References
- BioCRNpyler: Compiling chemical reaction networks from biomolecular parts in diverse contexts. William Poole, Ayush Pandey, Zoltan Tuza, Andrey Shur, Richard M. Murray. PLoS Computational Biology, 18(4), e1009987, 2022.
- Layered Feedback Control Overcomes Performance Trade-Off in Synthetic Biomolecular Networks. Chelsea Hu, Richard M. Murray. Nature Communications 13(1):1-13, 2022.
- Robustness Guarantees for Structured Model Reduction of Dynamical Systems. Ayush Pandey, Richard M. Murray. IEEE Conference on Decision and Control (CDC), 2021.
- Synthetic mammalian signaling circuits for robust cell population control. Yitong Ma, Mark W. Budde, Michaëlle N. Mayalu, Junqin Zhu, Richard M. Murray, Michael B. Elowitz. Submitted, Cell Systems.
- A two-state ribosome and protein model can robustly capture the chemical reaction dynamics of gene expression. Ayush Pandey, Richard M. Murray. 2020 Winter q-bio.
- Layered Feedback Control Improves Robust Functionality across Heterogeneous Cell Populations. Xinying Ren, Richard M. Murray. 2020 Winter q-bio.
- A geometric and structural approach to the analysis and design of biological circuit dynamics: a theory tailored for synthetic biology. John P. Marken, Fangzhou Xiao, Richard M. Murray. 2020 Winter q-bio.
- Theoretical Design of Paradoxical Signaling-Based Synthetic Population Control Circuit in E. coli. Michaëlle N. Mayalu, Richard M. Murray. 2020 Winter q-bio.
- Model Reduction Tools For Phenomenological Modeling of Input-Controlled Biological Circuits. Ayush Pandey, Richard M. Murray. 2020 Winter q-bio.
- An automated model reduction tool to guide the design and analysis of synthetic biological circuits. Ayush Pandey, Richard M. Murray. 2019 International Workshop on Biodesign Automation (IWBDA).
- Bacterial Controller Aided Wound Healing: A Case Study in Dynamical Population Controller Design. Leopold N. Green, Chelsea Y. Hu, Xinying Y. Ren, Richard M. Murray. qBio Conference 2019.
- Design of a genetic layered feedback controller in synthetic biological circuitry. Chelsea Y. Hu, Richard M. Murray. 2019 Synthetic Biology: Engineerinng, Evolution and Design (SEED) Conference.
- Control of density and composition in an engineered two-member bacterial community. Reed D. McCardell, Ayush Pandey, Richard M. Murray. Submitted, 2019 Synthetic Biology: Engineering, Evolution and Design (SEED) Conference.
- Hard Limits And Performance Tradeoffs In A Class Of Sequestration Feedback Systems. Noah Olsman, Ania-Ariadna Baetica, Fangzhou Xiao, Yoke Peng Leong, John Doyle, Richard Murray. Submitted, Cell Systems (Nov 2018).
- Guidelines for Designing the Antithetic Feedback Motif. Ania-Ariadna Baetica, Yoke Peng Leong, Richard M. Murray. Physical Biology, 17(5):055002, 2020.
- Cooperation Enhances Robustness of Coexistence in Spatially Structured Consortia. Xinying Ren, Richard M Murray. Submitted, 2019 European Control Conference.
- Model of Paradoxical Signaling Regulated T-Cell Population Control for Design of Synthetic Circuits. Michaelle N. Mayalu, Harman Mehta and Richard M. Murray. Submitted, 2019 European Control Conference (ECC).
- Role of interaction network topology in controlling microbial population in consortia. Xinying Ren, Richard M Murray. To appear, 2018 Conference on Decision and Control (CDC).
- Long-distance communication in synthetic bacterial consortia through active signal propagation. James M Parkin, Richard M Murray. 2018 Synthetic Biology: Engineering, Evolution and Design (SEED) Conference.
- Single day construction of multi-gene circuits with 3G assembly. Andrew D Halleran, Anandh Swaminathan, Richard M Murray. 2018 Synthetic Biology: Engineering, Evolution and Design (SEED) Conference.
- Control of bacterial population density with population feedback and molecular sequestration. Reed D McCardell, Shan Huang, Leopold N Green, Richard M Murray. 2018 Winter q-bio.
- Cell-free and in vivo characterization of Lux, Las, and Rpa quorum activation systems in E. coli. Andrew Halleran, Richard M. Murray. ACS Synthetic Biology, 7(2):752–755, 2017.
- Population regulation in microbial consortia using dual feedback control. Xinying Ren, Ania-Ariadna Baetica, Anandh Swaminathan, Richard M. Murray. Submitted, 2017 Conference on Decision and Control (CDC).
- Quantitative Modeling of Integrase Dynamics Using a Novel Python Toolbox for Parameter Inference in Synthetic Biology. Anandh Swaminathan, Victoria Hsiao, Richard M Murray. 2017 Synthetic Biology: Engineering, Evolution, and Design (SEED) Conference.
The project or effort depicted was or is sponsored by the Defense Advanced Research Projects Agency (Agreement HR0011-17-2-0008). The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
|
|
|