Biomolecular Circuits for Rapid Detection and Response to Environmental Events: Difference between revisions
No edit summary |
No edit summary |
||
| (20 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{righttoc}} This | {{righttoc}} This project funded by the [http://www.icb.ucsb.edu/ ARO Institute for Collaborative Biotechnologies], an Army University Affiliated Research Center (UARC) | ||
{| cellpadding=0 cellspacing=0 width=80% | {| cellpadding=0 cellspacing=0 width=80% | ||
| Line 5: | Line 5: | ||
| width=50% | | | width=50% | | ||
Current participants: | Current participants: | ||
{{project current participants}} | |||
Additional participants: | |||
{{project additional participants}} | |||
| | |||
<!-- Collaborators --> | |||
Previous participants: | |||
{{project previous participants}} | |||
* {{Emzo de los Santos}} | * {{Emzo de los Santos}} | ||
* {{Dan Siegal-Gaskins}}* | * {{Dan Siegal-Gaskins}}* | ||
|} | |} | ||
== Objectives == | == Objectives == | ||
[[Image:icb13-synbio.png| | [[Image:icb13-synbio.png|right|400px]] | ||
The goal of this project is to develop a set of biomolecular circuit modules for detecting molecular events that can be interconnected to create biological devices capable of monitoring the local environment around a cell, detecting and remembering complex temporal patterns, and triggering a response. We will build on [[Biomolecular Feedback Circuits for Modular, Robust and Rapid Response|previous ICB-supported work]] in design of biomolecular feedback circuits for modular, robust and rapid response, including design of proteins with programmable modulation of activity, design of domain-based scaffolds for programmable sensing and computa- tion, and development of forced response testing for signal response and robustness to environmental conditions. We will also exploit ongoing activities (funded by DARPA) in the development of biomolecular breadboards for proto- typing and debugging of biomolecular circuits. | The goal of this project is to develop a set of biomolecular circuit modules for detecting molecular events that can be interconnected to create biological devices capable of monitoring the local environment around a cell, detecting and remembering complex temporal patterns, and triggering a response. We will build on [[Biomolecular Feedback Circuits for Modular, Robust and Rapid Response|previous ICB-supported work]] in design of biomolecular feedback circuits for modular, robust and rapid response, including design of proteins with programmable modulation of activity, design of domain-based scaffolds for programmable sensing and computa- tion, and development of forced response testing for signal response and robustness to environmental conditions. We will also exploit ongoing activities (funded by DARPA) in the development of biomolecular breadboards for proto- typing and debugging of biomolecular circuits. | ||
| Line 25: | Line 26: | ||
== References == | == References == | ||
{{project paper list}} | |||
{{grant info| | |||
| agency = Army Research Office | |||
| grantno = | |||
| start = 1 Dec 2013 | |||
| end = 30 Nov 2018 | |||
| support = ~2 graduate students + supplies | |||
| reports= annual reports due in June | |||
| ID = ARO ICB | |||
}} | |||
[[Category:Completed projects]] | |||
[[Category:Biocircuits projects]] | |||
Revision as of 21:23, 26 May 2020
This project funded by the ARO Institute for Collaborative Biotechnologies, an Army University Affiliated Research Center (UARC)
|
Current participants: Additional participants: |
Previous participants:
|
Objectives
The goal of this project is to develop a set of biomolecular circuit modules for detecting molecular events that can be interconnected to create biological devices capable of monitoring the local environment around a cell, detecting and remembering complex temporal patterns, and triggering a response. We will build on previous ICB-supported work in design of biomolecular feedback circuits for modular, robust and rapid response, including design of proteins with programmable modulation of activity, design of domain-based scaffolds for programmable sensing and computa- tion, and development of forced response testing for signal response and robustness to environmental conditions. We will also exploit ongoing activities (funded by DARPA) in the development of biomolecular breadboards for proto- typing and debugging of biomolecular circuits.
Specific objectives for this project include:
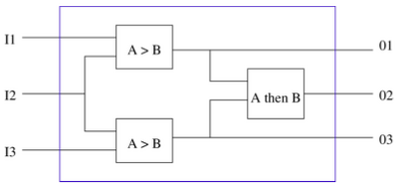
- Demonstrate individual components for signal detection, event memory, species comparison and basic logical operation in a mutually compatible set of technologies.
- Demonstrate a simple set of event detectors that trigger expression of a protein (reporter or enzyme) for the conditions “A > B” and “A followed by B”.
- Demonstrate the ability to interconnect individual event detectors to monitor the environment for more complex temporal patterns
References
- Control Theory for Synthetic Biology: Recent Advances in System Characterization, Control Design, and Controller Implementation for Synthetic Biology. Victoria Hsiao, Anandh Swaminathan, and Richard M. Murray. IEEE Control Systems Magazine, 38(3):32-62 , June 2018.
- Repressing Integrase attachment site operation with CRISPR-Cas9 in E. coli. Andrey Shur and Richard M Murray. Submitted, 2017 Synthetic Engineering: Engineering, Evolution and Design (SEED) Conference.
- Engineering pulsatile communication in bacterial consortia. James Parkin, Victoria Hsiao, Richard M Murray. Submitted, 2017 Synthetic Biology: Engineering, Evolution and Design (SEED) Conference.
- Engineering Transcriptional Regulator Effector Specificity Using Computational Design and In Vitro Rapid Prototyping: Developing a Vanillin Sensor. E. L. de los Santos, J. T. Meyerowitz, S. L. Mayo, R. M. Murray. ACS Synthetic Biology. 5(4):287-95, 2016.
- A population-based temporal logic gate for timing and recording chemical events. Victoria Hsiao, Yutaka Hori, Paul W.K. Rothemund, Richard M. Murray. Molecular Systems Biology, 12: 869, 2016.
- Design and application of stationary phase combinatorial promoters. Victoria Hsiao, Aileen Cheng and Richard M. Murray. Submitted, 2016 Synthetic Biology: Engineering, Evolution and Design (SEED) Conference.
- The role of single occupancy effects on integrase dynamics in a cell-free system. Georgios Artavanis, Victoria Hsiao, Clarmyra A. Hayes, Richard M. Murray. 2016 Synthetic Biology: Engineering, Evolution and Design (SEED) Conference (17 May 2016).
- Use of population-level cell-state switching for recording transient inducer pulses. Victoria Hsiao, Yutaka Hori, Paul W.K. Rothemund, Richard M. Murray. Presented, 2016 Winter q-Bio (5 Nov 2015).
- Engineering Transcriptional Regulator Effector Specificity using Computational Design and In Vitro Rapid Prototyping: Developing a Vanillin Sensor. Emmanuel Lorenzo Cornejo de los Santos, Joseph T Meyerowitz, Stephen L Mayo, Richard M Murray. ACS Synthetic Biology, 5(4):287–295, 2016..
- Characterization of minimum inducer separation time for a two-input integrase-based event detector. Victoria Hsiao, Yutaka Hori, and Richard M Murray. Presented, 2015 Winter q-bio Conference (5 Nov 2014).
- Engineering Transcriptional Regulator Effector Specificity Through Rational Design and Rapid Prototyping. Emmanuel L.C. de los Santos, Joseph T. Meyerowitz, Stephen L. Mayo, and Richard M. Murray. Presented at Synthetic Biology: Engineering, Evolution and Design (SEED), 14-18 July 2014.
- Negative autoregulation matches production and demand in synthetic transcriptional networks. Elisa Franco, Giulia Giordano, Per-Ola Forsberg, Richard M. Murray. Submitted, ACS Synthetic Biology (4 Oct 2013).
- Timing molecular motion and production with a synthetic transcriptional clock. Elisa Franco, Eike Friedrichs, Jongmin Kim, Ralf Jungmann, Richard Murray, Erik Winfree, Friedrich C. Simmel. Proceedings of the National Academy of Science (PNAS), 108 (40) E784-E793.
|
|
|