Theory-Based Engineering of Biomolecular Circuits in Living Cells
This is a joint AFOSR BRI project between MIT, Boston University, Caltech and Rutgers. This page primarily describes the work done in Richard Murray's group.
Project participants:
|
Collaborators:
|
Overview
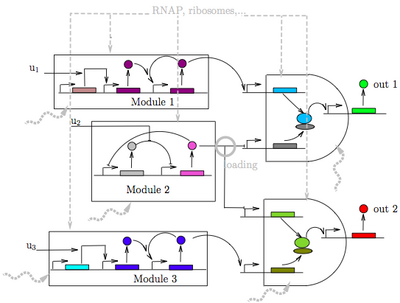
The objective of this research is to establish a data-driven theoretical framework based on mathematics to enable the robust design of interacting biomolecular circuits in living cells that perform complex decision making. Microbiology as a platform has substantial advantages with respect to human-made hardware, including size, power, and high sensitivity/selectivity. While the latest advances in synthetic biology have rendered the creation of simple functional circuits in microbes possible, our ability of composing circuits that behave as expected is still missing. This hinders the possibility of designing robust complex decision making, including recognition and classification of chemical signatures. Overcoming this bottleneck goes beyond the engineering of new parts or new assembly methods. By contrast, it requires a deep understanding of the dynamical interactions among synthetic modules and the cell machinery, a particularly hard task since dynamics are nonlinear, stochastic, and involve multiple scales of resolution both in time and space.
Project objectives:
- Establish a design-oriented theoretical framework that explicitly accounts for interactions among circuits, between the circuits and the cell machinery, and provides engineering solutions to mitigate the undesirable effects of these interactions;
- Develop design-oriented analysis tools to quantify the propagation of stochasticity through the nonlinear dynamics of biological networks;
- Develop a quantitative methodology to incorporate spatial heterogeneity effects into the analysis and design framework;
- Develop prototype experimental systems and a concrete demonstration of the integration of sensors that robustly distinguish between different chemical signatures.
Publications
- Data-driven network models for genetic circuits from time-series data with incomplete measurements. Enoch Yeung, Jim Kim, Ye Yuan, Jorge Gonçalves, Richard M. Murray. Journal of the Royal Society Interface 18 (182), 20210413.
- Analysis of primitive genetic interactions for the design of a genetic signal differentiator. Wolfgang Halter, Richard M. Murray, Frank Allgower. Submitted, OUP Synthetic Biology, May 2019.
- Quantitative characterization of random partitioning in the evolution of plasmid-encoded traits. Andrew D. Halleran, Emanuel Flores-Bautista, Richard M. Murray. qBio 2019 Conference.
- Cooperation Enhances Robustness of Coexistence in Spatially Structured Consortia. Xinying Ren, Richard M Murray. Submitted, 2019 European Control Conference.
- Control Theory for Synthetic Biology: Recent Advances in System Characterization, Control Design, and Controller Implementation for Synthetic Biology. Victoria Hsiao, Anandh Swaminathan, and Richard M. Murray. IEEE Control Systems Magazine, 38(3):32-62 , June 2018.
- Recursively constructing analytic expressions for equilibrium distributions of stochastic biochemical reaction networks. X. Flora Meng, Ania-Ariadna Baetica, Vipul Singhal, Richard M. Murray. Royal Society Interface, 14(130), 2017.
- Biophysical Constraints Arising from Compositional Context in Synthetic Gene Networks. Enoch Yeung, Aaron J. Dy, Kyle B. Martin, Andrew H. Ng, Domitilla Del Vecchio, James L. Beck, James J. Collins, Richard M. Murray. Cell Systems, 5(1):11–24.e12, 2017.
- Quantitative Modeling of Integrase Dynamics Using a Novel Python Toolbox for Parameter Inference in Synthetic Biology. Anandh Swaminathan, Victoria Hsiao, Richard M Murray. 2017 Synthetic Biology: Engineering, Evolution, and Design (SEED) Conference.
- A Bayesian approach to inferring chemical signal timing and amplitude in a temporal logic gate using the cell population distributional response. Ania-Ariadna Baetica, Thomas Anthony Catanach, Victoria Hsiao, Richard Murray, James Beck. 2017 Winter q-bio Conference.
- The Effect of Compositional Context on Synthetic Gene Networks. Enoch Yeung, Aaron J Dy, Kyle B Martin, Andrew H Ng, Domitilla Del Vecchio, James L Beck, James J. Collins, Richard M Murray. Submitted, Cell Systems (bioRxiv preprint).
- Stochastic Gene Expression in Single Gene Oscillator Variants. Anandh Swaminathan, Marcella M. Gomez, David L. Shis, Matthew R. Bennett, and Richard M. Murray. 2016 Synthetic Biology: Engineering, Evolution and Design (SEED) Conference.
- Finding stationary solutions to the chemical master equation by gluing state spaces at one or two states recursively. Xianglin Meng, Ania A. Baetica, Vipul Singhal, and Richard M. Murray. Presented, 2016 Winter q-Bio Conference.
- Rapid in vitro engineering of 16 two-input logic gates. Clarmyra A. Hayes, Emmanuel L.C. de los Santos, Enoch Yeung, Sean R. Sanchez, Seung Y. Lee, and Richard M. Murray. Presented, 2016 Winter q-Bio.
- Linear System Identifiability from Distributional and Time Series Data. Anandh Swaminathan and Richard M. Murray. 2016 American Control Conference (ACC).
- A stochastic framework for the design of transient and steady state behavior of biochemical reaction networks. Ania A. Baetica, Ye Yuan, Jorge Goncalves and Richard M. Murray. 2015 Conference on Decision and Control (CDC).
Research supported by the Air Force Office of Scientific Research, grant number FA9550-14-1-0060.
|
|
|