Establishing microfluidic cell-free systems for the rapid prototyping of synthetic genetic networks
This is a joint project with Sebastian Maerkl at EPFL in which we propose to combine our expertise in microfluidics, synthetic biology, and computational modeling to implement existing in vivo systems in vitro and vice versa. This study is designed to demonstrate that biological systems can be forward and reverse engineered in cell-free environments.
|
Project participants:
|
Collaborators:
|
Objectives
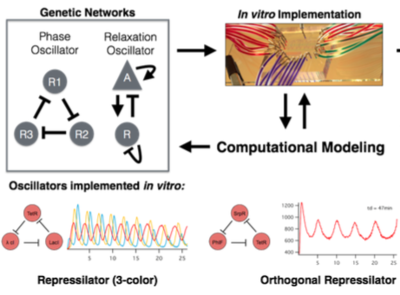
Computational modeling is instrumental to guiding the development of any genetic system. In vitro implementation of genetic networks allows tuning of numerous parameters, many not accessible in vivo such as dilution rates and DNA template concentrations. Computational models allow experimentalists to efficiently traverse a smaller space of possible parameter combinations leading to the successful implementation of in vitro and in vivo synthetic networks. We will develop computational models for the three oscillators (two in vivo, one in vitro) studied here. These models will provide initial guidelines on how to implement existing oscillators in vitro and insights into why certain genetic oscillators are robust in vitro whereas others are not. To further improve characterization and optimization of genetic networks in vitro we will develop control algorithms capable of fully automating a microfluidic platform to: i) automatically determine system parameters such as transcription/translation rates, repression/activation rates, etc. and ii) efficiently traverse the parameter space of complex genetic regulatory networks in vitro. We propose to develop a closed feedback system that controls the microfluidic system, runs experiments and analyses results to automatically redefine the parameter sets in the next round of experiments.
References
- Modeling Dynamic Transcriptional Circuits with CRISPRi. Samuel E Clamons, Richard M Murray. 2018 Winter q-bio.
Research supported by the Human Frontiers Science Program.
|
|
|