An Open Synthetic Biology Toolkit for Engineering Reliable Genetic Circuits in Microbes in Soil
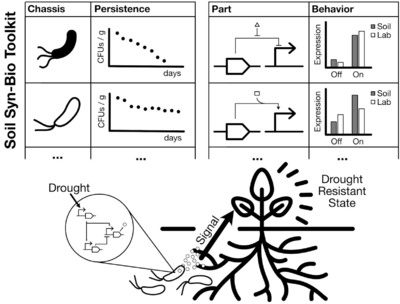
This goal of this project is to demonstrate the feasibility of an “open source” toolkit for soil synthetic biology, with the goal of bootstrapping a larger effort that would enable the use of engineered microbes to understand and modulate the complex dynamics of the rhizosphere. We will identify and characterize genetically tractable microbes capable of long-term persistence; create a toolbox of genetic parts for gene circuits and pathways in soil conditions; and design, build, and test a stimulus-response circuit operating in soil. Long-term applications include engineering microbial communities to optimize nutrient uptake and improve survival against environmental hazards such as drought, toxins, or pathogens.
|
Current participants: Additional participants: |
Collaborators:
Past participants:
|
Objectives
The extensive progress that synthetic biology has made in the past 20 years on engineering microbes to perform complex tasks such as logical computation, non-native metabolic pathways, and autonomous control functions makes the field well-poised to address the sustainability challenges facing our society today, e.g. by increasing crop yield by providing plants with the ability to dynamically respond to conditions like drought or disease. However, soil synthetic biology presents a unique set of challenges that have yet to be addressed substantially by the field. An engineered microbial system must be able to persist and maintain its function within a predictable range in the soil over months- or even years-long timescales. As the developments in synthetic biology have thus far been implemented primarily in E. coli cells in exponential-phase growth in laboratory conditions, it is unlikely that they can be directly applied in situ to a soil-based application. Our project will address this need for a well-characterized toolkit of strains and genetic parts with which to build biological circuits that can function reliably in long-term soil environments. Our specific objectives are to:
- Identify & characterize genetically-tractable microbes capable of long-term soil persistence
- Create a toolbox of genetic parts for building gene circuits for long-term soil deployment
- Design, build, and test a stimulus-response circuit with predictable function in soil
References
- Metabolic engineering of Pseudomonas putida for production of vanillylamine from lignin-derived substrates. João Heitor Colombelli Manfrão-Netto, Fredrik Lund, Nina Muratovska, Elin M. Larsson, Nádia Skorupa Parachin, Magnus Carlquist. Microbial Biotechnology, 14( 6), 2448– 2462, 2021.
This work was supported by the Resnick Sustainability Institute.
|
|
|