Engineering Durable Cell-Free Biological Capabilities for Advanced Sensing and Prototyping
The goal of this project is to systematically expand the operating range and utility of cell-free systems sourced from new diverse organisms. These next-generation cell free systems will enable new capabilities for prototyping and implementing durable synthetic circuit designs. Such standardized systems will not only explore the boundaries of cell-free prototyping and characterization but will also enable proper comparisons of data measured by multiple groups, thereby accelerating the usage and broader utility of cell-free systems in industry and academia.
|
Current participants: Additional participants: |
Collaborators: Past participants:
|
Objectives
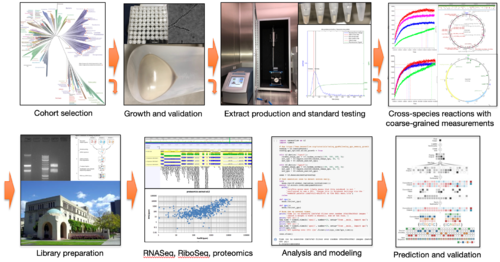
The two primary focus areas for this project are to (1) increase the number of organisms and portable parts available for synthetic biologists for design, prototyping, and implementation of cell-free biomolecular circuits and pathways in a rapid, systematic, and predictable way and (2) improve the durability, sensitivity, and robustness of cell-free detection systems. The specific objectives for the project are:
- Develop high-throughput, cell-free methods for measuring and modeling key parameters of transcriptional and translational processes of biological parts in a diverse set of organisms.
- Perform a demonstration of the utility of these models by moving parts and circuits from one organism to another with a repeatable and predictable outcome.
- Demonstrate expanded capabilities for detection and diagnostics of chemical and biological signals in simulated or real field operating conditions, with the goal of increasing sensitivity, specificity, and output signal strength of cell-free biological detectors.
- Distribute the tools and techniques to the synthetic biology community for broader use.
References
- Development of cell-free transcription-translation systems in three soil Pseudomonads. Joseph T. Meyerowitz, Elin M. Larsson, Richard M. Murray. To appear, ACS Synthetic Biology, 2024.
- A Method for Cost-Effective and Rapid Characterization of Genetic Parts. John B. McManus, Casey B. Bernhards, Caitlin E. Sharpes, David C. Garcia, Stephanie D. Cole, Richard M. Murray, Peter A. Emanuel, Matthew W. Lux. To appear, Journal of Visualized Experiments (JoVE).
- A Method for Cost-Effective and Rapid Characterization of Engineered T7-based Transcription Factors by Cell-Free Protein Synthesis Reveals Insights into the Regulation of T7 RNA Polymerase-Driven Expression. John B. McManus, Richard M. Murray, Peter A. Emanuel, Matthew W. Lux. Archives of Biochemistry and Biophysics, Volume 674, 15 October 2019, 108045.
This research is supported by the Institute for Collaborative Biotechnologies through contract W911NF-19-D-0001 from the U.S. Army Research Office. The content of the information on this page does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
|
|
|