Biomolecular Breadboards for Prototyping and Debugging Synthetic Biocircuits
This is a joint project between Richard Murray (Caltech), Vincent Noireaux (U. Minnesota) and Paul Rothemund (Caltech), funded by the DARPA Living Foundries program. The information on this page focuses primarily on the work involving my research group. Additional information about this project is available on our OpenWetWare web pages.
Project participants:
|
Collaborators
|
* Partially supported
Objectives
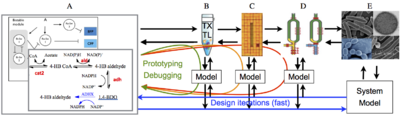
|
| Overview of the cell-free expression breadboard process. |
The goal of this project is to build a set of “biomolecular breadboards” to create a systematic, engineering-oriented approach to synthesizing biomolecular circuits that involves developing, modeling, and debugging a sequence of prototype devices, each at increasing levels of complexity and each allowing the incorporation of increasingly realistic operating environments for either in vitro or in vivo applications. We are adapting an existing cell-free toolbox developed at U. Minnesota to create a set of prototyping environments for testing biological circuits. A sequence of increasingly complex environments, ending in prokaryotic cells, will be used to demonstrate the ability to prototype circuits that function in vivo, with iteration in in vitro assays and models to streamline development of predictable, in vivo functionality.
Phase I (complete)
- Post protocols for producing basic cell-free breadboard on public web site along with summary of costs. Provide sequences for positive controls.
- Demonstrate the use of cell-free breadboard on 2 existing circuits (chosen from negatively autoregulated gene, simple genetic switch, oscillator, feedforward loop) and document time required to implement a simple circuit and perform design iterations.
- Post complete documentation of a transcription-translation cell-free platform that includes a set of transcriptional repression units (non-degradable and degradable versions), the working principles of the toolbox (procedures and protocols), and validated models for the repressors based on in vitro and in vivo characterization.
- Demonstrate the design of a simple circuit (3–6 unique promoters) that consists of a new set of genetic elements, documenting the interactions, conditions, and compensation mechanisms required to obtain a working circuit in E. coli. Investigators shall provide an analysis of the design cycle times compared to iGEM standard assembly protocol and Gibson- based protocol (using in vivo testing and debugging). Target 3 day prototyping cycle time and 1 month from design to implementation, with the ability to test 25 circuit variants simultaneously.
Phase II
- Train interested researchers in the use of the cell-free breadboard system, including a design project that demonstrates the design, modeling, implementation and debugging of a simple circuit
- Demonstrate the ability to use a combination of modeling combined with a sequence of in vitro design cycles to get a novel circuit (4–8 unique promoters) working in E. coli.
- Demonstrate the ability to get a modest complexity circuit (8–16 unique promoters, based on composing simpler circuits) working in E. coli. Target 1 day prototyping cycle time and 1 week from design to implementation, with the ability to test 100 circuit variants simultaneously.
- Develop, document and dessiminate a set of protocols and technologies enabling the use of digital microfluidics (droplets) to carry out TX-TL reactions for prototyping and debugging of biological circuits.
- Demonstrate the ability to create and screen circuit combinations using high throughput ($10^6$ droplets/day) microfluidics, demonstrating 2-4X improvement in pathway yield.
References
- Quantifying Resource Competition and its Effects in the TX-TL System. Andras Gyorgy and Richard M. Murray. 2016 Conference on Decision and Control (CDC).
- Rapid cell-free forward engineering of novel genetic ring oscillators. Henrike Niederholtmeyer, Zachary Sun, Yutaka Hori, Enoch Yeung, Amanda Verpoorte, Richard M Murray and Sebastian J Maerkl. eLife 2015;10.7554/eLife.09771.
- Characterizing and Prototyping Genetic Networks with Cell-Free Transcription-Translation Reactions. Melissa K Takahashi, Clarmyra A. Hayes, James Chappell, Zachary Z. Sun, Richard M Murray, Vincent Noireaux, Julius B. Lucks. Methods, 15(85):60-72, 2015.
- Engineering Principles of Synthetic Biochemical Oscillators with Negative Cyclic Feedback. Yutaka Hori and Richard M. Murray. Submitted, 2015 Conference on Decision and Control (CDC).
- A State-space Realization Approach to Set Identification of Biochemical Kinetic Parameters. Yutaka Hori and Richard M. Murray. 2015 European Control Conference (ECC).
- Synthetic logic circuits using RNA aptamer against T7 RNA polymerase. Jongmin Kim, Juan F Quijano, Enoch Yeung, Richard M Murray. bioRxiv, 4 Sep 2014.
- Protein degradation in a TX-TL cell-free expression system using ClpXP protease. Zachary Z. Sun, Jongmin Kim, Vipul Singhal, Richard M. Murray. Technical Report, 7 July 2014.
- System identification of phosophorylation based insulator in a cell-free in vitro transcription-translation system. Enoch Yeung, Shaobin Guo, Richard M. Murray. Submitted, 2014 Winter q-bio Conference (5 Nov 2013).
- Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. Zachary Z. Sun, Enoch Yeung, Clarmyra A. Hayes, Vincent Noireaux, Richard M. Murray. Submitted, ACS Synthetic Biology, September 2013.
- Biomolecular resource utilization in elementary cell-free gene circuits. Dan Siegal-Gaskins, Vincent Noireaux, and Richard M. Murray. 2013 American Control Conference (ACC).
- DARPA Living Foundries program review presentation, 11 Feb 2014
- An In Silico Modeling Toolbox for Rapid Prototyping of Circuits in a Biomolecular “Breadboard” System, Zoltan A. Tuza, Vipul Singhal, Jongmin Kim, Richard M. Murray. Submitted, 2013 Conference on Decision and Control (CDC).
- Biomolecular resource utilization in elementary cell-free gene circuits, D. Siegal-Gaskins, V. Noireaux, and R. M. Murray. American Control Conference (ACC), 2013.
- Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology, Z. Z. Sun, C. A. Hayes, J. Shin, F. Caschera, R. M. Murray and V. Noireaux. Journal of Visualized Experiments (JoVE), 2013.
- OpenWetWare Biomolecular Breadboards project page
- DARPA Living Foundries kickoff presentation, 12 Jul 2012
Research supported by the Defense Advanced Research Projects Agency (DARPA/MTO) Living Foundries program, contract number HR0011-12-C-0065. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing official policies, either expressly or implied, of the Defense Advanced Research Projects Agency or the U.S. Government.
|
|
|