Theory-Based Engineering of Biomolecular Circuits in Living Cells: Difference between revisions
(Created page with "<hr> {{grant info| | agency = Air Force Office of Scientific Research | grantno = | start = 1 Oct 2013 | end = 30 Sep 2018 | support = ~2 graduate students + supplies | repor...") |
No edit summary |
||
| Line 1: | Line 1: | ||
{{righttoc}} | |||
This is a joint project between MIT, Boston University, Caltech and Rutgers. This page primarily describes the work done in Richard Murray's group. | |||
{| width=80% | |||
|- valign=top | |||
| width=50% | Current participants: | |||
* TBD | |||
| width=50% | Collaborators: | |||
* Domitilla Del Vecchio (MIT) | |||
* Jim Collins (BU) | |||
* Eduardo Sontag (Rutgers) | |||
Past participants: | |||
|} | |||
== Overview == | |||
[[Image:afosr13-synbio.png|right|400px]] | |||
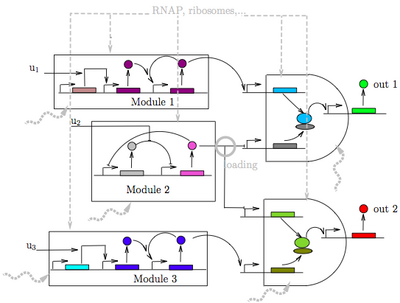
The objective of this research is to establish a data-driven theoretical framework based on mathematics to enable the robust design of interacting biomolecular circuits in living cells that perform complex decision making. Microbiology as a platform has substantial advantages with respect to human-made hardware, including size, power, and high sensitivity/selectivity. While the latest advances in synthetic biology have rendered the creation of simple functional circuits in microbes possible, our ability of composing circuits that behave as expected is still missing. This hinders the possibility of designing robust complex decision making, including recognition and classification of chemical signatures. Overcoming this bottleneck goes beyond the engineering of new parts or new assembly methods. By contrast, it requires a deep understanding of the dynamical interactions among synthetic modules and the cell machinery, a particularly hard task since dynamics are nonlinear, stochastic, and involve multiple scales of resolution both in time and space. | |||
== Publications == | |||
None to date. | |||
<hr> | <hr> | ||
{{grant info| | {{grant info| | ||
Revision as of 18:09, 28 September 2013
This is a joint project between MIT, Boston University, Caltech and Rutgers. This page primarily describes the work done in Richard Murray's group.
Current participants:
|
Collaborators:
Past participants: |
Overview
The objective of this research is to establish a data-driven theoretical framework based on mathematics to enable the robust design of interacting biomolecular circuits in living cells that perform complex decision making. Microbiology as a platform has substantial advantages with respect to human-made hardware, including size, power, and high sensitivity/selectivity. While the latest advances in synthetic biology have rendered the creation of simple functional circuits in microbes possible, our ability of composing circuits that behave as expected is still missing. This hinders the possibility of designing robust complex decision making, including recognition and classification of chemical signatures. Overcoming this bottleneck goes beyond the engineering of new parts or new assembly methods. By contrast, it requires a deep understanding of the dynamical interactions among synthetic modules and the cell machinery, a particularly hard task since dynamics are nonlinear, stochastic, and involve multiple scales of resolution both in time and space.
Publications
None to date.
|
|
|