Networked Feedback Systems in Biology: Difference between revisions
From Murray Wiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| (28 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{righttoc}} This is a joint project with [http://www.cds.caltech.edu/~doyle John Doyle], funded by the [http://www.icb.ucsb.edu/ ARO Institute for Collaborative | {{righttoc}} This is a joint project with [http://www.cds.caltech.edu/~doyle John Doyle], funded by the [http://www.icb.ucsb.edu/ ARO Institute for Collaborative Biotechnologies]. This page primarily describes the work done in Richard Murray's group. | ||
{| cellpadding=0 cellspacing=0 | {| cellpadding=0 cellspacing=0 width=80% | ||
|- valign=top | |- valign=top | ||
| width=50% | | | width=50% | | ||
Current participants: | Current participants: | ||
* {{ | * {{Vanessa Jonsson}} | ||
* {{ | * {{Ophelia Venturelli}} | ||
* {{Enoch Yeung}} | |||
* {{Dan Siegal-Gaskins}} | |||
| | | | ||
Previous participants: | Previous participants: | ||
* Dionysios Barmpoutis | |||
* Mary Dunlop | |||
* Per-Ola Forsberg | * Per-Ola Forsberg | ||
* Elisa Franco | |||
* Roman Galeev | * Roman Galeev | ||
* | | | ||
* | <br> | ||
* Arthur Prindle | |||
* Henrik Sandberg | |||
* Johan Ugander | |||
* Kevin Welch | |||
|} | |} | ||
== Objectives == | == Objectives == | ||
{| style="float: right" width=40% border=1 | |||
|- | |||
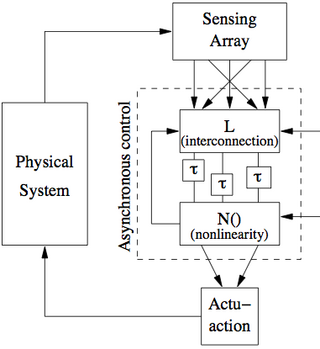
| [[Image:biomolecular-feedback.png|320px]]<br> | |||
Candidate architecture for biomolecular control system. The feedback system consists of the process dynamics, an arrange of molecular sensing systems, an "networked" control system and a collection of actuation mechanisms. The networked control system makes use of an interconnection matrix <amsmath>L</amsmath>, a set of asynchronous delays (represented by the blocks | |||
labeled <amsmath>\tau</amsmath>) and nonlinear elements <amsmath>N(\cdot)</amsmath>. Internal feedback between the nonlinear block and the interconnection block allows a general set of dynamical systems to be formed from this simple structure. | |||
|} | |||
This project explores the application of theoretical approaches to complex systems in the domain of biological networks. Our focus is twofold: on the development and application of analytical tools for the analysis and design of complex networks, and on the use of those techniques to construct novel biological circuits for feedback control of cellular processes. Specific activites include: | |||
We are developing a | * ''Design of programmable, RNA-based platforms for control in biological networks''. We are exploring the use of RNA-based feedback circuits for regulation of biosynthetic pathways and developing methods that allow larger scale networks to be constructed in a modular fashion. | ||
* ''Modularity and composition of biological circuits and subsystems''. In order to build useful models for the design of large-scale biological networks, it will be necessary to combine models of smaller subnetworks that interact with each other. While the theory for such interconnection is well developed in physical and information systems, such a theory is not present in biological systems, where coupling between the indi-vidual circuits is present through a variety of mechanisms. | |||
* ''Analysis and design tools for complex networked systems, driven by biological circuit design''. Biology integrates computation, communications, control, thermodynamics and statistical mechanics in a way that does not allow current theories to be applied beyond small subsystems. Tools that can be used to understand and design biological control systems will require new methods that capture robustness, fragility, evolvability and modularity in new ways. We believe that new theoretical approaches, driven by our in-terets in biological circuit design, can provide useful tools for understanding large-scale, complex networks in a variety of application areas. | |||
== Publications == | == Publications == | ||
* [http://www.cds.caltech.edu/~murray/papers/2013f_ybm13-cdc.html Modeling Environmental Disturbances with the Chemical Master Equation], Enoch Yeung, James L. Beck, and Richard M. Murray. Submitted, 2013 Conference on Decision and Control (CDC). | |||
* [http://www.cds.caltech.edu/~murray/papers/2013b_jmm13-cdc.html Reverse Engineering Combination Therapies for Evolutionary Dynamics of Disease: An Hinfty Approach], Vanessa Jonsson, Nikolai Matni and Richard M. Murray. Submitted, 2013 Conference on Decison and Control (CDC). | |||
* [http://www.cds.caltech.edu/~murray/papers/2012h_yeu+12-cdc.html Quantifying Crosstalk in Biochemical Systems], Enoch Yeung, Jongmin Kim, Ye Yuan, Jorge Goncalves and Richard M. Murray. 2012 Conference on Decision and Control (CDC). | |||
* [http://www.cds.caltech.edu/~murray/papers/2012y_vem12-pnas.html Synergistic dual positive feedback loops established by molecular sequestration generate robust bimodal response], Ophelia S. Venturellia, Hana El-Samad and Richard M. Murray. Proceedings of the National Academy of Science (PNAS), 2012. | |||
* {{fdm09-cdc}} | |||
* {{Dun_08-natgen}} | |||
* M. J. Dunlop, E. Franco, R. M. Murray. "A Multi-Model Approach to Identification of Biosynthetic Pathways." In Proceedings of the 26th American Control Conference 2007. | |||
* {{FM08-cdc}} | |||
* E. Franco, P. O. Forsberg and R. M. Murray. "Design, modeling and synthesis of an in vitro transcription rate regulatory circuit". American Control Conference, 2008. | |||
* J. Ugander, M. J. Dunlop, R. M. Murray. "Analysis of a Digital Clock for Molecular Computing." In Proceedings of the 26th American Control Conference 2007. | |||
* {{JU08-ms}} | |||
== Miscellaneous == | |||
* [http://openwetware.org/wiki/BioControl BioControl Journal Club] | |||
[[Category:Completed projects]] | |||
{{#set: agency=ARO ICB | end date = 2013}} | |||
Latest revision as of 21:23, 26 May 2020
This is a joint project with John Doyle, funded by the ARO Institute for Collaborative Biotechnologies. This page primarily describes the work done in Richard Murray's group.
|
Current participants:
|
Previous participants:
|
|
Objectives
This project explores the application of theoretical approaches to complex systems in the domain of biological networks. Our focus is twofold: on the development and application of analytical tools for the analysis and design of complex networks, and on the use of those techniques to construct novel biological circuits for feedback control of cellular processes. Specific activites include:
- Design of programmable, RNA-based platforms for control in biological networks. We are exploring the use of RNA-based feedback circuits for regulation of biosynthetic pathways and developing methods that allow larger scale networks to be constructed in a modular fashion.
- Modularity and composition of biological circuits and subsystems. In order to build useful models for the design of large-scale biological networks, it will be necessary to combine models of smaller subnetworks that interact with each other. While the theory for such interconnection is well developed in physical and information systems, such a theory is not present in biological systems, where coupling between the indi-vidual circuits is present through a variety of mechanisms.
- Analysis and design tools for complex networked systems, driven by biological circuit design. Biology integrates computation, communications, control, thermodynamics and statistical mechanics in a way that does not allow current theories to be applied beyond small subsystems. Tools that can be used to understand and design biological control systems will require new methods that capture robustness, fragility, evolvability and modularity in new ways. We believe that new theoretical approaches, driven by our in-terets in biological circuit design, can provide useful tools for understanding large-scale, complex networks in a variety of application areas.
Publications
- Modeling Environmental Disturbances with the Chemical Master Equation, Enoch Yeung, James L. Beck, and Richard M. Murray. Submitted, 2013 Conference on Decision and Control (CDC).
- Reverse Engineering Combination Therapies for Evolutionary Dynamics of Disease: An Hinfty Approach, Vanessa Jonsson, Nikolai Matni and Richard M. Murray. Submitted, 2013 Conference on Decison and Control (CDC).
- Quantifying Crosstalk in Biochemical Systems, Enoch Yeung, Jongmin Kim, Ye Yuan, Jorge Goncalves and Richard M. Murray. 2012 Conference on Decision and Control (CDC).
- Synergistic dual positive feedback loops established by molecular sequestration generate robust bimodal response, Ophelia S. Venturellia, Hana El-Samad and Richard M. Murray. Proceedings of the National Academy of Science (PNAS), 2012.
- Design of insulating devices for in vitro synthetic circuits, Elisa Franco, Domitilla Del Del Vecchio, Richard M Murray. Conference on Decision and Control (CDC), 2009.
- Mary J Dunlop, Robert Sidney Cox, Joseph H Levine, Richard M Murray and Michael B Elowitz, Regulatory activity revealed by dynamic correlations in gene expression noise. Nature Genetics, 40:1493-1498, 2008.
- M. J. Dunlop, E. Franco, R. M. Murray. "A Multi-Model Approach to Identification of Biosynthetic Pathways." In Proceedings of the 26th American Control Conference 2007.
- Elisa Franco and Richard M Murray, Design and performance of in vitro transcription rate regulatory circuit. Conference on Decision and Control (CDC), 2008.
- E. Franco, P. O. Forsberg and R. M. Murray. "Design, modeling and synthesis of an in vitro transcription rate regulatory circuit". American Control Conference, 2008.
- J. Ugander, M. J. Dunlop, R. M. Murray. "Analysis of a Digital Clock for Molecular Computing." In Proceedings of the 26th American Control Conference 2007.
- J. Ugander, Delay-dependent Stability of Genetic Regulatory Networks. Masters thesis, Department of Automatic Control, Lund University, 2008.