TX-TL projects, 2015-16
|
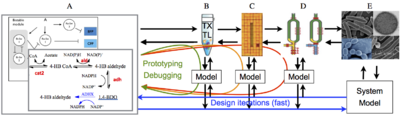
| Overview of the cell-free expression breadboard process. |
The overall goal of this research is to build a set of “biomolecular breadboards” to create a systematic, engineering-oriented approach to synthesizing biomolecular circuits that involves developing, modeling, and debugging a sequence of prototype devices, each at increasing levels of complexity and each allowing the incorporation of increasingly realistic operating environments for either in vitro or in vivo applications. We are adapting an existing cell-free toolbox developed at U. Minnesota to create a set of prototyping environments for testing biological circuits. A sequence of increasingly complex environments, ending in prokaryotic cells, is being used to demonstrate the ability to prototype circuits that function in vivo, with iteration in in vitro assays and models to streamline development of predictable, in vivo functionality.
A number of projects are available for 2015-16 for undergraduates interested in contributing to the research progress in this area. Possible projects include:
- Variability in circuit performance across extract preparation methods. We have seen many situations in which circuits work only in batches of extract produced with certain methods of lysis (bead beating, homogenization, french press, etc). There are no obvious patterns of what types of circuits work in what types of extract, and so there are a lot of interesting speculation about what is going on. Work in this area would require testing existing circuits in different extract batches and then debugging the circuits in the batches where they don't work correctly.
- Expression of membrane-bound proteins in TX-TL. Many circuit components require membrane bound proteins, including ligand-binding proteins and light-responsive transcription factors. Several of these components have been tested in TX-TL but have not yet shown activity. In addition, in metabolic engineering there are several membrane bound components of core metabolic pathways that are currently not accessible for TX-TL based prototyping. There are several promising approaches for integrating membrane-like structures into cell-free environments (). We will characterize several of these approaches and demonstrate one or more that can be used to express functional, membrane-associated proteins.
- Implementation of DNA editing machinery in TX-TL. Many groups are using DNA editing machinery as part of biological circuits. The two most common are CRISPR/Cas9 and integrase/excisionase systems. The Murray lab has demonstrated that some integrase/excisionase systems are functional while others are not, and has also performed initial (so far unsuccessful) testing of CRISPR/Cas 9. We plan to develop the protocols required to implement CRISPR-based circuits as well as integrase-based circuits, as well as explore the ability of these circuits to work on linear DNA (useful for rapid prototyping) versus plasmid DNA.
- Implementation of scaffold-based circuits using non-homologous kinases in TX-TL. The Murray lab has built several circuits that make use of scaffold-based kinases to implement feedback regulation strategies (Hsiao et al 2013). Repeated attempts at Caltech to implement these circuits in TX-TL have so far failed to create working circuits. By making use of the improved extracts in WP1 along with more sophisticated analysis techniques (eg, micro-arrays, fluorescent probes and fluorophore/quencher systems), we will debug these circuits and enable them to be used as components in TX-TL circuits.
More information on TX-TL is available here: http://www.openwetware.org/wiki/Biomolecular_Breadboards