Center for Harnessing Microbiota from Military Environments (CHARMME): Difference between revisions
No edit summary |
No edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 26: | Line 26: | ||
{{project paper list}} | {{project paper list}} | ||
[[Category: | [[Category:Active projects]] | ||
[[Category:Biocircuits projects]] | |||
{{Project | {{Project | ||
|Title=Center for Harnessing Microbiota from Military Environments (CHARMME) | |Title=Center for Harnessing Microbiota from Military Environments (CHARMME) | ||
| Line 37: | Line 38: | ||
|Reporting requirements=Quarterly reports | |Reporting requirements=Quarterly reports | ||
|Project ID=ARO CHARMME | |Project ID=ARO CHARMME | ||
|ack= | |ack=Research was sponsored by the Army Research Office and was accomplished under Cooperative Agreement Number W911NF-22-2-0210. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Office or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for Government purposes notwithstanding any copyright notation herein. | ||
}} | }} | ||
Revision as of 16:50, 17 March 2023
The Center for Harnessing Microbiota from Military Environments (CHARMME) is a multi-university, basic research project focused on predictive design of engineered cellular systems in defined environments.
|
Current participants:
Additional participants: |
Collaborators:
Past participants: |
Objectives
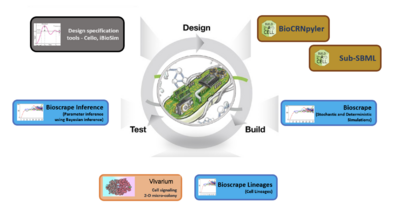
Caltech's efforts on focused on development of tools from control and dynamical systems to provide design rules and a computational framework that allows designers to assess robustness of their designs and to evaluate compensation mechanisms designed to enhance robustness. These techniques will build on existing modeling and design tools (BioCRNpyler, Cello, iBioSim, and others), but will integrate representations of biological context to allow distributions of responses to be computed as used as a means of assessing whether a device or circuit will function robustly across chassis and environmental contexts.
References
- Metabolic perturbations to an E. coli-based cell-free system reveal a trade-off between transcription and translation. Manisha Kapasiawala and Richard M. Murray. SEED 2023.
Research was sponsored by the Army Research Office and was accomplished under Cooperative Agreement Number W911NF-22-2-0210. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Office or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for Government purposes notwithstanding any copyright notation herein.
|
|
|