SURF 2024: Towards a Minimal Model for Virus-Host Interactions
2024 SURF project description
- Mentor: Richard Murray
- Co-mentors: Zachary Martinez
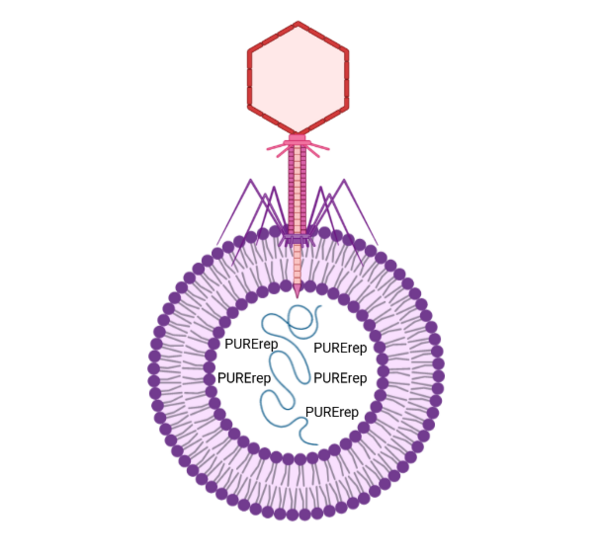
Main Idea: Engineering a synthetic cell that can be infected by a natural virus.
Introduction
Viruses are abundant (~1031), ancient (> 4 bya), and evolve quickly (average of 10-3 - 10-8 mutation rate). Due to the immense diversity of viruses, between 60-95% of viruses are uncharacterized/unidentifiable.1,2,3 Viruses can have global impacts including carbon cycling in the oceans and the spread of deadly disease during a pandemic. While most may consider viruses to be completely detrimental to humanity, they have proven to be incredibly useful for biotechnological purposes. From tailored phage therapy that can fight antibiotic resistant bacterial infections to the therapeutic delivery of base editors to correct genetic mutations like hemophilia, viruses can be powerful tools.4 While viruses are undoubtedly important to science, they can be extremely challenging to study. Complex lifestyles, sheer diversity, host requirement, and numerous other factors contribute to the hurdles that contemporary virology faces. Through the bottom-up construction of a synthetic host, we can hopefully exert greater control over our experimental design and allow for a more in-depth interrogation of the underlying viral biology. At the same time, with the advancement of synthetic biology, it would be useful to create a viral vector that can engage with synthetic cells. By allowing viruses to interface with synthetic cells, it opens up more complex engineering goals that might involve targeted delivery of genetic payloads (CRISPR/Cas9) to mixed populations of living and synthetic cells. The specificity and robustness of viruses might prove to be a valuable asset in our synthetic cell toolbox.
Project Overview
In order to build a minimal model for viral infection, we will be using synthetic cells for our hosts and T4 phage for our virus. Our non-living synthetic cells are typically comprised of cell-free protein synthesis extract that is encapsulated inside of a lipid vesicle. For this minimal model however, we will be embedding E. coli protein OmpC into our liposome in order for T4 to recognize the vesicle as a potential host. While we will first use RNA based toehold switches to determine if successful infection occurs, the end goal is to use PURErep (platform for transcription-translation-coupled DNA replication) inside of the vesicles which should allow for viral propagation after infection.5 Lastly, we will investigate whether the viruses are able to escape their host and what mechanism they use to lyse the liposome. If we are able to fully recapitulate the viral life cycle in a synthetic host, this will hopefully lead to a publication.
Preferred Skills
- Background in basic molecular biology through coursework
- Some experimental experience through coursework with a lab component or research
References
- Microbiology by numbers. Nat Rev Microbiol. 2011 Aug. https://doi.org/10.1038/nrmicro2644
- Nasir A., et al. Investigating the Concept and Origin of Viruses. 2020 Dec. https://doi.org/10.1016/j.tim.2020.08.003
- Stern A. and Andino R. Viral Evolution. Viral Pathogenesis. 2016 Feb. https://doi.org/10.1016%2FB978-0-12-800964-2.00017-3
- Rosen S., et al. Activity of transgene-produced B-domain-deleted factor VIII in human plasma following AAV5 gene therapy. Blood. 2020 Nov. https://doi.org/10.1182/blood.2020005683
- Libicher K., et al. In vitro self-replication and multicistronic expression of large synthetic genomes. Nature Communications. 2020 Feb. https://doi.org/10.1038/s41467-020-14694-2