SURF 2024: Establish synthetic biology toolkits for Steinernema nematode transgene expression
SURF 2024 project description
- Mentor: Mengyi Cao
- If you are interested in this topic, please check out our twin project (co-mentored by Elin Larsson):
Project Overview:
Steinernema nematodes associate with mutualistic symbiotic bacteria Xenorhabdus in a species-specific manner, and together they can infect insects that are agricultural pests. Therefore, the nematode-bacteria partnership is a valuable experimental system to study symbiosis and is important in promoting agricultural productivity. Although they were discovered a century ago, the molecular genetic tools in Steinernema nematodes are still limiting. Currently there is no consistent transgenesis protocol in these organisms, which severely restricted the visualization and control of gene expression.
Here are a few rate-limiting steps in developing such technique:
- Seek for suitable promoters that are native to the Steinernema species.
- Optimize the combination of regulatory elements.
- Choose the efficient selection/screen markers.
- Reduce the labor for transgene delivery.
Recently, synthetic biology toolkits, such as a part library, is proven to be a powerful system in designing various combinations of bacterial genetic elements which rapidly optimized the multigene biocircuit constructions (1–3). If similar synthetic biology pipeline can be adapted to multi-cellular eukaryotic organisms, such as nematodes, it will speed up transgenesis tool development and enable the study of ubiquitous and tissue-specific gene expression. In this project, we propose to establish a part library in S. hermaphroditum (India), a recently isolated nematode strain with a complete genome and is available for basic CRISPR-Cas9 genome editing (4,5).
Specific Aims:
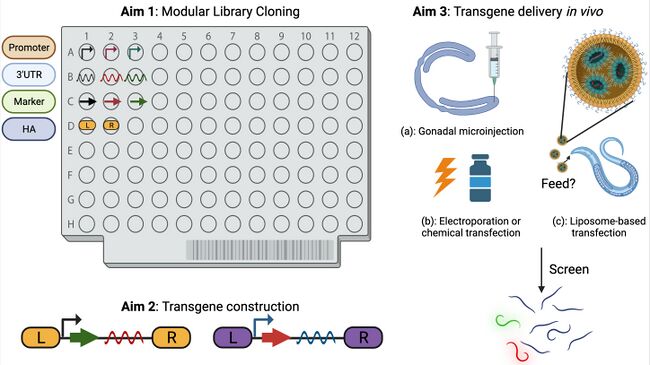
Aim 1: Cloning of modular parts.
We will construct a small-scale library by first cloning modular parts, including (1a): regulatory elements such as various lengths of promoters, 3’UTR sequences, and the first intron; (1b): screen or selection markers, such as codon-optimized fluorescence proteins and hygromycin-resistance cassette; and (1c): ‘inserting sequences’ including homology arms for CRISPR-Cas9, or ITR for transposon piggyBac, which will be used to insert the constructs into the targeted sites in S. hermaphroditum genome. The modular part library will be stored in E. coli and organized in 96-wells plates.
Aim 2: Transgene part library construction.
We will use a Golden Gate-Gibson (3G) cloning pipeline to construct combinations of the modular parts (from Aim 1) and build a library.
Aim 3: Transgene delivery in vivo.
We will deliver these constructs via gonadal microinjection, either based on CRISPR-Cas9, extrachromosomal array, or transposition. We will also attempt a pilot experiment using a liposome-based transfection and electroporation. The latter two delivery approaches, if successful, will significantly relieve the labor from microinjection, and speed up the molecular genetics tool development in Steinernema and potentially other nematode species.
This proposed effort will be the first step to build comprehensive bioengineering toolkits that will be crucial to tissue-specific gene expression visualization, quantification, and manipulation. If successful at a small scale, the transgene part library can be expanded to a larger scale to build other tools, such as tissue-specific CRISPR, Gal4-UAS gene manipulation, and optogenetics tools.
Preferred Candidate: (fulfills one or more of the following)
- Previous experience with molecular cloning
- Bioengineering-oriented
- Interested in nematode-bacteria symbiosis system
- Basic knowledge in genetics
References:
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A Modular Cloning System for Standardized Assembly of Multigene Constructs. PLoS ONE. 2011;6(2):e16765.
- Iverson SV, Haddock TL, Beal J, Densmore DM. CIDAR MoClo: Improved MoClo Assembly Standard and New E. coli Part Library Enable Rapid Combinatorial Design for Synthetic and Traditional Biology. ACS Synth Biol. 2016;5(1):99–103.
- Halleran AD, Swaminathan A, Murray RM. Single Day Construction of Multigene Circuits with 3G Assembly. Acs Synth Biol. 2018;7(5):1477–80.
- Cao M. CRISPR-Cas9 genome editing in Steinernema entomopathogenic nematodes. bioRxiv. 2023;2023.11.24.568619.
- Cao M, Schwartz HT, Tan CH, Sternberg PW. The entomopathogenic nematode Steinernema hermaphroditum is a self-fertilizing hermaphrodite and a genetically tractable system for the study of parasitic and mutualistic symbiosis. Genetics. 2021;220(1).