SURF 2021: Optimizing cell extract for a proto-flagellar system
SURF 2021 project description
- Mentor: Richard Murray
- Co-mentor: Manisha Kapasiawala
Introduction
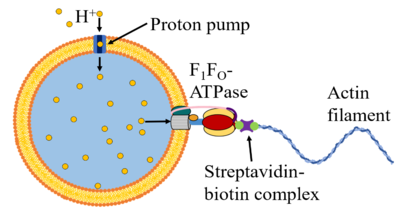
Past efforts in synthetic biology have created significant advancements towards the creation of synthetic cells [1]. In the simplest case, synthetic cells are phospholipid vesicles encapsulating a solution of cell extract and energy buffer, which provide the chemical components (e.g. RNA polymerase, ATP, NTPs, amino acids, etc.) for transcription and translation (TX-TL) and basic metabolism [2,3]. As minimal chassis, however, synthetic cells provide a powerful platform for both understanding natural cell behaviors and for programming synthetic behaviors, as the bottom-up construction of these synthetic cells from fully characterized components enables predictability and functionality through mechanistic insight. Recent work has shown that synthetic cells can be programmed to perform behaviors that are observed in natural cells, such as to deliver cargo, process signals from environmental inputs, and make complex decisions in response to those signals. In the Murray Lab, we are currently focusing on the development of motility in synthetic cells, by repurposing the F1FO-ATP synthase (ATPase) rotary motor to construct an ATPase-based protoflagellum (see figure). Here, proton pumps in the synthetic cell membrane will create a proton gradient that will drive the rotation of ATPase, which will in turn drive the rotation of an actin filament that will propel the synthetic cell.
Currently, the method of introducing the proton pump and ATPase to the synthetic cell membrane is a laborious and expensive process that involves purifying these proteins from E. coli and reconstituting them in synthetic cells. An easier way to reconstitute these proteins in synthetic cells would be to have them make these proteins themselves, but doing so remains challenging due to experimental limitations.
Research overview
The SURF student will use computational modeling to help optimize cell extract for a proto-flagellar system, by creating a modeling framework for protein production that captures cell extract metabolism. At the most basic level, protein production in cells is modeled as two reactions: transcription and translation. Each reaction can be described mathematically, and the resulting chemical reaction network (CRN) can be simulated to predict protein production (e.g. how much protein is made, how long it takes to reach that steady state level, etc.). Starting from a basic CRN, one can build increasing complex CRNs by considering that there are finite amounts of ribosomes and RNA polymerases in a cell, considering dilution and degradation of mRNA and proteins, etc.
Protein production in cell extract, however, is very different from protein production in a cell, largely because there is no recycling of metabolic products and of energy molecules such as ATP and NADH. Thus, in cell extract, the metabolic reactions going on “in the background”, which we can typically ignore when modeling protein production in a cell, may have a significant impact on protein production, whether via depletion of ATP and other resources, toxic accumulation of phosphate, pH effects, etc. Creating protein production models that capture the effects of cell extract metabolism may lead to the creation of more accurate protein production models.
The SURF project will focus on the creation of a modeling framework for protein production that will capture the effects of cell extract metabolism. To create this modeling framework, the student will use BioCRNpyler [4], a tool developed by the Murray Lab to quickly and easily build CRNs, to model the core metabolic reactions occurring in cell extract. Incorporating transcription and translation reactions will result in the creation of a model where protein production is coupled to cell extract metabolism.
Using parameters derived from literature and previous experiments done in the lab, the SURF student will create and compare models with metabolism vs. those without to determine the effects of cell metabolism on protein production. As far as a specific focus for this project goes, there are two possible directions that a SURF student can pursue as applications of this modeling framework.
Creating design specifications for in vesicle production of the protoflagellar system
As a proof-of-concept for the modeling framework described above, the student will first work on a protein production model for bacteriorhodopsin (proton pump) and ATP synthase, two components that are necessary for the proto-flagellar system. If sufficient progress is made, using insights from this model, the student can also work experimentally towards the goal of achieving and improving protein production of bacteriorhodopsin and ATP synthase, both in bulk extract and in encapsulated vesicles. Questions that the student may pursue include the following: how much ATP is needed for the production of sufficient bacteriorhodopsins and ATP synthases? does changing the starting pH of the cell extract improve protein production? how does the starting concentration of NADH/NADPH/other molecules impact protein production in this system?
Creating guidelines for metabolic engineering of cell-free protein synthesis
Cell extract contains a lot of enzymes that are useful for E. coli, but not necessarily useful for cell-free protein production. These enzymes participate in reactions that divert resources like ATP and amino acids away from protein production and towards pathways that are unproductive (one study found that only 12% of energy resources in cell extract went towards protein production!). An interesting long-term goal would be to create a cell line where the enzymes that participate in these unproductive pathways have been experimentally knocked out, so that when we make cell extract, we will have fewer resources diverted away from protein production. However, knocking out important pathways may result in E. coli that are not viable. This project will aim to computationally "knock out" enzymes in their cell-free protein production model, while modeling the effects of these knockouts in living cells, to determine the optimal metabolic engineering strategy for improving protein production. While similar work has been explored in other labs, the work has either been experimental work unguided by mathematical intuition, or theoretical work that fails to consider possible experimental limitations [5, 6, 7]. If successful, this project will help pave the way for future experimental work in creating this optimized cell extract.
SURF student qualifications:
- Prerequisite coursework: Bi1x (or similar introductory biology course that provides a basic foundation in molecular biology)
- Preferred coursework: BE 150, BE/APh 161
- Other: basic familiarity with coding preferred (Python, Java, etc.)
References:
- P. Stano, “Is Research on ‘Synthetic Cells’ Moving to the Next Level?,” Life, vol. 9, no. 1, 2019, doi: 10.3390/life9010003.
- N. E. Gregorio, M. Z. Levine, and J. P. Oza, “A User’s Guide to Cell-Free Protein Synthesis,” Methods and Protocols, vol. 2, no. 1, 2019, doi: 10.3390/mps2010024.
- H. Jia, M. Heymann, F. Bernhard, P. Schwille, and L. Kai, “Cell-free protein synthesis in micro compartments: building a minimal cell from biobricks,” New Biotechnology, vol. 39, pp. 199–205, Oct. 2017, doi: 10.1016/j.nbt.2017.06.014.
- W. Poole, A. Pandey, A. Shur, Z. A. Tuza, and R. M. Murray, “BioCRNpyler: Compiling Chemical Reaction Networks from Biomolecular Parts in Diverse Contexts,” bioRxiv, p. 2020.08.02.233478, Jan. 2020, doi: 10.1101/2020.08.02.233478.
- N. Horvath, M. Vilkhovoy, J. A. Wayman, K. Calhoun, J. Swartz, and J. D. Varner, “Toward a genome scale sequence specific dynamic model of cell-free protein synthesis in Escherichia coli,” Metabolic Engineering Communications, vol. 10, p. e00113, Jun. 2020, doi: 10.1016/j.mec.2019.e00113.
- R. W. Martin et al., “Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids,” Nature Communications, vol. 9, no. 1, p. 1203, Mar. 2018, doi: 10.1038/s41467-018-03469-5.
- H. J. Lim and D.-M. Kim, “Cell-Free Metabolic Engineering: Recent Developments and Future Prospects,” Methods and Protocols, vol. 2, no. 2, 2019, doi: 10.3390/mps2020033.