SURF 2020: System identification of bacterial gene expression as a function of population dynamics
2020 SURF: project description
- Mentor: Richard M. Murray
- Co-mentor: Chelsea Hu
Gene expression is a complex process that is often described by a coarse-grained model with only a few states and a handful of parameters [1]. For instance, if we consider a coarse-grained deterministic model that describes the transcriptional and translational dynamics of a gene encoding fluorescent protein P and its mRNA M in the equation. box:
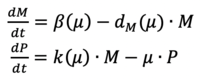
Where μ represents the growth rate of the cell population, which is also the dominant effect in protein dilution and degradation. Additionally, β and dM denote the transcription and degradation rate of mRNA, respectively; k is the translation rate of protein. In coarse-grained models, we often assume constant cell division rate in controlled cell culture conditions. Therefore, β, dM, and k are estimated to be constant parameters. These assumptions allow parameters to become identifiable and therefore provide useful insights on a system’s dynamics. However, They do not always hold when synthetic organisms are deployed to a real-world environment. Specifically, there has been a large amount of both theoretical and experimental work, which shows that gene expression is growth rate dependent [2,3,4]. In the equations above, the parameters β, dM, and k are no longer constants but functions of growth rate μ. Multiple environmental factors could contribute to a growth rate change, including nutrient availability, metabolic burdens, temperature, pH levels, and the ecological context. It is, therefore, unclear whether cell growth has a causal effect on gene expression or if gene expression is merely subject to growth conditions.
The objective of this study is to determine the topology and parameters of cellular gene expression as a function of bacterial population dynamics. To achieve this goal, deterministic and stochastic system identification methods [5,6] will be used along with experimentally collected molecular and population dynamics. This study is a combination of theoretical/ computational and experimental work. Skills and interests in both molecular cloning and programming are desired.
References
[1] Alon, U. An Introduction to Systems Biology. Chapter 2. (Chapman and Hall/CRC, 2006). doi:10.1201/9781420011432
[2] Klumpp, S. & Hwa, T. Bacterial growth: global effects on gene expression, growth feedback and proteome partition. Curr. Opin. Biotechnol. 28, 96–102 (2014).
[3] Klumpp, S., Zhang, Z. & Hwa, T. Growth Rate-Dependent Global Effects on Gene Expression in Bacteria. Cell 139, 1366–1375 (2009).
[4] Friedman, J. & Gore, J. Ecological systems biology: The dynamics of interacting populations. Current Opinion in Systems Biology 1, 114–121 (2017).
[5] Swaminathan, A., Poole, W., Hsiao, V. & Murray, R. M. Fast and flexible simulation and parameter estimation for synthetic biology using bioscrape. bioRxiv 54, 2548–27 (2019).
[6] Hu, C. Y., Varner, J. D. & Lucks, J. B. Generating Effective Models and Parameters for RNA Genetic Circuits. ACS Synth Biol 4, 914–926 (2015).