SURF 2019: Evolutionary Stability of Genetic Circuits
2019 SURF project description
- Mentor: Richard Murray
- Co-mentor: Andy Halleran
Evolutionary Stability of Genetic Circuits
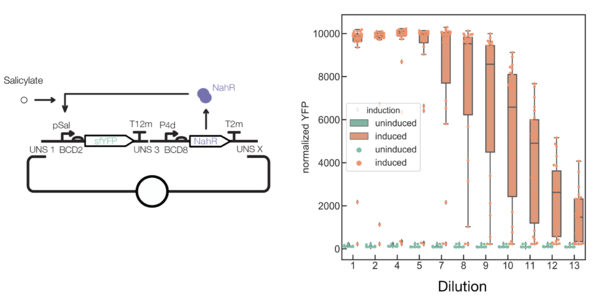
Synthetic biology’s heralded inception promised revolutions in practically every field, from agriculture and manufacturing to medicine. However, all synthetic biology faces an uphill battle: synthetic circuits require host resources, inevitably slowing host growth. Therefore, mutants that disable the synthetic circuit grow faster than cells faithfully carrying out the programmed function. This causes mutants to outcompete engineered cells, bringing an end to the synthetic program (Figure 1).
Figure 1: Cells containing an inducible activation circuit where addition of the small molecule salicylate drives sfYFP expression decrease in fluorescence over time.
This universal feature of synthetic biology imposes a limit on the lifetime of programmed cellular functions. Improving the lifetime of synthetic circuits is of fundamental importance to the field of synthetic biology. The goal of this SURF project is to improve our understanding of the evolutionary stability of genetic circuits. This is a general aim and there are many potential projects that range from more biology focused (i.e. understanding the biology underpinning the stability of synthetic circuits) to more engineering focused (i.e. building circuits to improve lifetime). A few example projects are given below.
1) Genetic circuit architecture and evolutionary
- Compare different molecular implementations for the same desired behavior and see if there are differences in stability (e.g. repressor vs. activator for inducible transcription)
2) Cellular response to synthetic burden
- We have preliminary data that shows once burdened cells go into stationary phase they have difficulty resuming exponential growth when they are transferred to fresh media. What's causing this physiological change, and can we characterize how burden forces this response?
3) Link expression of burdensome genes to ribosomal protein level
- Previous work by Alon and Dekel (Shachrai et al., 2010, Mol Cell) demonstrated that synthetic protein expression is more costly when the cell has fewer ribosomes. This project would aim to link expression of synthetic circuit to the level of ribosomal proteins in the cell to minimize cellular burden.
Useful previous experience
There are no requirements for previous experience. The following could be useful depending on the project.
- molecular biology / cloning (PCR, bacterial transformation, culturing + miniprep, etc.)
- time-lapse microscopy
- image analysis
- statistics
- coding (python / matlab)
- mathematical modeling (ODEs, deterministic / stochastic simulations)
References