SURF 2018: Modeling the Effect of Intracellular Signaling Mechanisms on Population Dynamic Behaviors in the Context of Paradoxical Signaling
2018 SURF: project description
- Mentor: Richard M. Murray
- Co-mentor: Michaëlle N. Mayalu
Description
Background
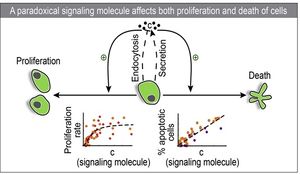
Cells can communicate with each other by secreting signaling molecules that diffuse between them. Cell detection of an extracellular signaling molecule triggers cascades of intracellular signaling events that regulate diverse cell behaviors such as the proliferation, apoptosis, differentiation, and gene expression. In some cases, the exact same signaling molecule can affect opposing or antagonistic cell behaviors. This scenario is referred to as paradoxical signaling. Two important examples of paradoxical signaling are:
1. Beta cells secrete insulin, which lowers blood glucose levels. Glucose has both mitogenic and toxic effects on the cell1.
2. T cells secret a cytokine IL-2 which promotes T cell proliferation2 and also affects cell death3.
Cells can use paradoxical signaling to balance between proliferation and death rates and it is therefore an effective method to regulate homeostasis of cell population. By modeling these paradoxical signaling systems we can gain insight into how defects in population control affect diseases such as cancer, type II diabetes and autoimmunity4. In addition, synthetic biology has begun to exploit paradoxical signaling mechanisms to engineer new cell population control circuits.
Although cellular behavior and cell-cell communication are regulated by the internal biochemical signaling mechanisms within individual cells, connecting these mechanisms to the overall population dynamics poses a considerable challenge for multiple reasons including:
1. Complexity and/or limited knowledge of intracellular mechanisms
2. Differences in length and time scales between molecular-level reactions and cell-level dynamics
Goal
The goal of this project is to develop and explore different modeling techniques that can describe how intracellular mechanisms lead to overall cell population behavior in the context of paradoxical signaling. Possible tasks include (but are not limited to):
1. Extending, modifying and/or combining modeling frameworks found in literature
2. Developing reasonable justifications for modeling simplifications and assumptions
3. Conducting model parameter studies and drawing insights into system behavior
Recommended Skills:
1. Systems Biology
2. Strong familiarity in Linear and Nonlinear Differential Equations
3. Basic Linear Algebra
4. Statistics
5. Proficiency in Matlab and or Python
References:
2. Zhu, J. & Paul, W. E. CD4 T cells: fates, functions, and faults. Blood 112, 1557–1569 (2008).
6. Youk, H. & Lim, W. A. Sending Mixed Messages for Cell Population Control. Cell 158, 973–975 (2014).