SURF 2018: Integrase-based genetic circuits
2018 SURF project description
- Mentor: Richard Murray
- Co-mentor: Andrey Shur
Integrases are bacteriophage proteins that catalyze recombination of phage DNA with the bacterial genome at specific DNA sequences known as attachment (att) sites. Serine integrases in particular catalyze directional recombination between cognate att sites without additional cofactors and therefore are valuable tools in synthetic biology. Previous work has utilized serine integrases and attachment sites as black boxes that inducibly flip or excise DNA to construct genetic memory, logic gates, and event detectors. We recognize that the ability to induce DNA changes can be a powerful tool in synthetic biology, since DNA is an easily tractable medium for information storage and of course encodes all the proteins and genes needed for life. Thus we are interested in expanding the list of different "functions" that integrases can perform as well as thinking about new and interesting ways to utilize the functions that are already known. A few descriptions of ongoing projects in this area are below.
Integrase-based continuous event logging
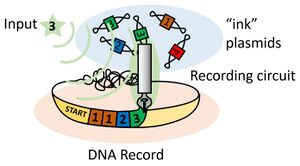
Biological records of events are omnipresent in paleontology, history, and climate science. Tree rings and ice cores provide evidence of environmental conditions that have been recorded in the composition of living cells that are deposited over time, carrying with them the a record of events that have influenced their lives before being buried underneath ice or inside the trunk of a tree. Using integrases, customizable biological memory devices can be made to record any event that a bacterium can detect. Bacteria can be unobtrusively seeded in a natural environment, providing a stable genetic record on any timescale that is suitable to the researcher.
Field programmable circuits in bacteria
Microchips with a specific purpose are expensive and time consuming to make. For many applications, it is sufficient to make a chip containing a set of unconnected logic gates and other elements, and allow the user to define the connections between these components after the chip has already been made. This concept is known as a programmable interconnect. Using integrases it is possible to develop a programmable interconnect in bacteria. A set of circuits and useful components can be integrated in the bacterial genome in a non-functional form. This way, the genes are not impacting the health of the bacterium if they are not needed. A set of integrase operations can then be used to selectively enable and connect disparate circuit elements in vivo, without the need for traditional mo-lecular cloning. This could allow one strain to have many different uses and functions without compromising the viability and stability of its genome. Users would simply program the strain before use, removing or not activating unneeded circuit components at will.
Prerequisite Skills
- Molecular cloning
- Golden Gate assembly
- Gibson assembly
- Restriction cloning
- Primer design
- Bacteria culture
- Miniprep
- Python
References
- Bonnet, J., Subsoontorn, P. & Endy, D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. 109, 8884–8889 (2012).
- Roquet, N., Soleimany, A. P., Ferris, A. C., Aaronson, S. & Lu, T. K. Synthetic recombinase-based state machines in living cells. Science (80-. ). 353, aad8559-aad8559 (2016).
- Hsiao, V., Hori, Y., Rothemund, P. W. K. & Murray, R. M. A population-based temporal logic gate for timing and recording chemical events. Mol. Syst. Biol. 557, 1–17 (2016).
- Smith, M. C. A., Till, R. & Smith, M. C. M. Switching the polarity of a bacteriophage integration system. Mol. Microbiol. 51, 1719–1728 (2004).
- Shur, A. & Murray, R. M. Repressing Integrase attachment site operation with CRISPR-Cas9 in E. coli. bioRxiv (2017). doi:10.1101/110254
- Shur, A. & Murray, R. M. Proof of concept continuous event logging in living cells. bioRxiv (2017). doi:10.1101/225151