SURF 2014: Rapid prototyping of moderate complexity biomolecular circuits
2014 SURF project description
- Mentor: Richard M. Murray
- Co-mentor: Clare Hayes
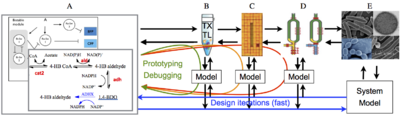
|
| Overview of the cell-free expression breadboard process. |
The overall goal of this project is to build a set of “biomolecular breadboards” to create a systematic, engineering-oriented approach to synthesizing biomolecular circuits that involves developing, modeling, and debugging a sequence of prototype devices, each at increasing levels of complexity and each allowing the incorporation of increasingly realistic operating environments for either in vitro or in vivo applications. We are adapting an existing cell-free toolbox developed at U. Minnesota to create a set of prototyping environments for testing biological circuits. A sequence of increasingly complex environments, ending in prokaryotic cells, is being used to demonstrate the ability to prototype circuits that function in vivo, with iteration in in vitro assays and models to streamline development of predictable, in vivo functionality.
This SURF project will focus on the design and implementation of a modest complexity (8-12 promoter) circuit using the TX-TL breadboard that has been previous developed. Students will select a circuit during the SURF proposal process. During the summer, the design will proceed in several (partially overlapping) phases:
- The circuit will be modeled using an existing TX-TL toolbox (built on top of MATLAB) and this model will be used to verify the basic dynamics of the circuit, as well as used to test hypotheses as the circuit is implemented and debugged.
- Individual components of the circuit will be built and tested using the TX-TL experimental breadboard. Experimental iterations typically take 1-2 days per iteration, with up to 50 variations tested per cycle. Measurements will be taken using fluorescent reporters and a plate reader.
- Once component operation is verified (experimentally and using mathematical models), the components will be composed in the TX-TL toolbox and adjusted as needed to demonstrate functionality.
- Components, combinations of components and the full system will also be tested in vivo, but transforming the circuits into lab strains of E. coli and measured using either a plate reader or a flow cytometer.
Required skills: Students interested in this project should have some basic laboratory skills (equivalent to Bi 1X, Bi 10 or ChE/BE 130) and have experience with either MATLAB, Python or a similar scientific programming package.
References
- An In Silico Modeling Toolbox for Rapid Prototyping of Circuits in a Biomolecular “Breadboard” System, Zoltan A. Tuza, Vipul Singhal, Jongmin Kim, Richard M. Murray. Submitted, 2013 Conference on Decision and Control (CDC).
- Biomolecular resource utilization in elementary cell-free gene circuits, D. Siegal-Gaskins, V. Noireaux, and R. M. Murray. American Control Conference (ACC), 2013.
- Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology, Z. Z. Sun, C. A. Hayes, J. Shin, F. Caschera, R. M. Murray and V. Noireaux. Journal of Visualized Experiments (JoVE), 2013.
- OpenWetWare Biomolecular Breadboards project page
- DARPA Living Foundries kickoff presentation, 12 Jul 2012