SURF 2014: Fabricating a Toolbox of RNA Thermometers
2014 SURF project description
- Mentor: Richard Murray
- Co-mentor: Shaunak Sen
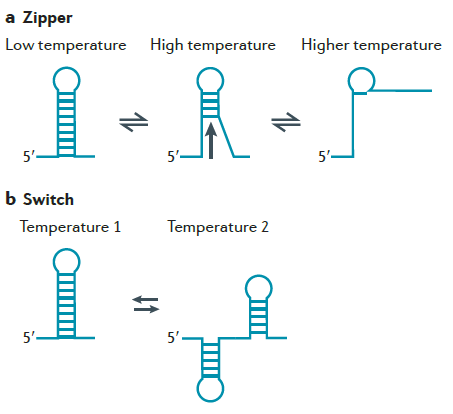
Overview: RNA Thermometers (RNATs) are mRNA sequences that change their secondary structures on the basis of temperature. Natural RNATs are usually present upstream of genes and regulate their rate of translation by revealing or hiding the Ribosome Binding Site (RBS) at different temperatures. Two common types of RNATs [4] are:
a) Switch-type: These RNATs ‘switch’ between two mutually exclusive secondary structures at different temperatures. One example is the cIII switch found in λ phage viruses.
b) Zipper-like: These are simple stem-loop structures which melt at higher temperatures and re-anneal at lower temperature. Examples include the ROSE (Repression Of heat Shock gene Expression) elements in many bacteria.
Previous studies [1] [3] have shown how simple ‘zipper-like’ RNATs can be synthesized de novo. It is possible to alter the structures of these RNATs and observe temperature sensitivity of gene expression with respect to these changes. I will be working on developing a toolkit of RNATs with different threshold values of gene expression in relation to changes in temperature. Further, I will be demonstrating the utility of our toolkit through a simple gene circuit.
Goals:
1. Validating putative synthetic RNATs [1] in vitro using the Transcription-Translation toolkit (TXTL) [5] and coming up with new designs based on free energy estimation using Mfold [2] as well as testing them in vitro
2. Testing these novel RNAT designs in vivo
3. Application of the toolkit in construction of a gene circuit that gives a temperature-compensated output for a given input
Required Skills: This project requires experience in molecular biology techniques and cloning. Also, understanding of as well as interest in synthetic biology and computation modeling is a pre-requisite.
References
[1] J. Neupert, D. Karcher, R. Bock. “Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli.” Nucleic Acids Res. (2008): 36(19).
[2] http://mfold.rna.albany.edu/?q=mfold (accessed date: 05-30-14).
[3] J. Neupert, R. Bock. "Designing and using synthetic RNA thermometers for temperature-controlled gene expression in bacteria." Nature protocols 4.9 (2009): 1262-1273.
[4] J. Kortmann, F. Narberhaus. “Bacterial RNA thermometers: molecular zippers and switches.” Nature Reviews Microbiology 10 (2012): 255-265.
[5] ZZ. Sun, CA. Hayes, J. Shin, F. Caschera, RM. Murray, V. Noireaux. "Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology." JoVE (Journal of Visualized Experiments) 79 (2013): e50762-e50762.