Genetic Circuits for Multi-Cellular Machines
The use of microbial consortia for implementing synthetic circuits and biosynthesis pathways has a number of advantages over design using single strains, including separation and specialization of func- tion, reduction of loading on individual cells, and reuse of limited molecular and genetic components. However, differences in growth rate between different organisms in the consortium and the effects of mutation on community function can interfere with consortium function. We propose an experimental framework for distributing circuit and pathway functionality across a collection of cells, and regulating the effects of differential growth rate and mutation in microbial consortia.
Project participants:
|
Collaborators:
|
Objectives
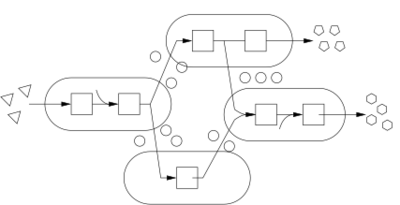
Year 1: In the first year of the project we will develop the basic components that are required to implement genetic circuits for multi-cellular machines:
- Demonstrate 2-strain synchronization to cell-cell signaling pulse, triggered by external “stress” event (temperature, pH, etc).
- Demonstrate the ability to utilize integrase-mediated recombination to "differentiate" bacteria by simultaneously activating circuit/pathway expression and ceasing plasmid replication.
- Model and experimentally validate the dynamics of 2-4 species communities exchanging metabolites and responding to cell-cell signaling, both in well-mixed and spatially structured conditions.
Year 2: In the second year of the project, we will integrate the individual signaling and regulation func- tions and demonstrate the ability to implement multi-cellular machines capable of implementing distributed circuits and pathways:
- Combine metabolic cross-feeding community structures with cell-signaling circuits and growth control circuits, and demonstrate the ability to maintain stable community structures that respond to environmental inputs/stress factors.
- Utilize engineered consortia with differentiation/conditional plasmid replication as chassis for multi-step complex biosynthesis of compounds of Army interest (e.g., terpenoids, flavonoids, alkaloids, or polyketides). Compare yield, robustness, and duration of production with/without differentiation and metabolic cross-feeding structures.
References
- Length and time scales of cell-cell signaling circuits in agar. Joy Doong, James Parkin, Richard M. Murray. 2018 Winter q-bio.
This research is supported by the Institute for Collaborative Biotechnologies through grant W911NF-09-D-0001 from the U.S. Army Research Office. The content of the information on this page does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
|
|
|