SURF 2021: Modeling tools for design and analysis of synthetic biological circuits
SURF 2021 project description
- Mentor: Richard Murray
- Co-mentor: Ayush Pandey
Introduction:
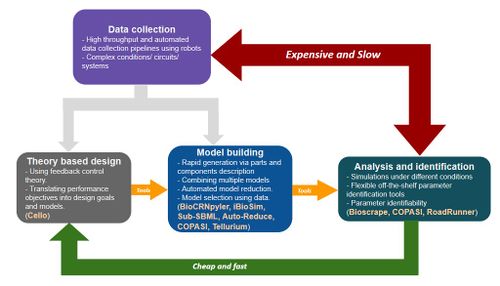
This SURF project is about modeling, simulations, and analysis methods and tools used in engineering biological circuits. Figure 1 shows a brief overview of the pipeline starting at the design phase where biological controllers are designed to be implemented either in vivo or in vitro to achieve certain biological / chemical function. To specify the performance specifications, to predict signalling levels, and to analyze these biological circuits, mathematical models are built. Control theoretic methods [1] can be used to study the stability and performance using these mathematical models. Similar to other engineering disciplines, experimental data is used to identify the model parameters in order to make design decisions. However, unlike other engineering disciplines there are various challenges such as biological complexity, context dependence, and lack of understanding of various interactions in a biological system that obscure the use of models as the predictive design tools. Hence, this is an active area of research [2] with various interesting directions of research as suggested in Figure 1.
Research overview:
A 10-12 week project on modeling and analysis tools for biological circuits is possible in various research directions. As shown in Figure 1, modeling plays an important role in the standard design-build-test cycle for synthetic biology. This role can be divided into three parts as shown in the figure viz. theory based design of biological circuits, building and selecting models that represent a given circuit, and finally using experimental data to identify parameters of the models and study the related properties using simulations and other analysis tools. Each of these is discussed briefly in the following bullets:
- Biological feedback controllers can be designed at molecular and/or at cell population level. Control theory principles and analysis tools for stability and performance are commonly used to study the various properties that can be expected from a circuit and also to assess new design ideas. For example, in order to design a genetic oscillator, it is important to study the multi stability properties of the proposed nonlinear circuit models. Similarly, we have been exploring the question of studying how these properties arise from the particular nonlinear structure of chemical reaction network models.
- From the parts and components description of a biological circuit, creating models that represent the circuit functions and the dynamics is an important task. Automated tools such as TX-TLsim [3] and iBioSim [4] can be used to create chemical reaction network models of a circuit. We have been working on developing a similar Python based chemical reaction network compiler called BioCRNpyler [6]. It can be used to quickly create models for biological circuits given the parts, components, and mechanism description. The models of all the submodules can then be assembled together by other tools such as Sub-SBML [7]. BioCRNpyler is primarily aimed at creating models for cell-free systems but can also be used to create subsystem models of circuits in vivo. We are working on developing these tools further and using them to model and simulate synthetic cell vesicles and cell-free circuits.
- To validate and quantify the models for a circuit, experimental data is used to identify the model parameters. Often for biological systems a big challenge is that the output measurements cannot be used to identify all the model parameters uniquely. A set of parameter identification tools is available in a Cython based fast stochastic simulator toolbox called bioscrape [5]. Various kinds of data from different experiments can be used to validate these tools and the identified models can be used to study system properties of these circuits. Moreover, signalling levels can be predicted and used for circuit improvements and design with the identified models.
Research directions for the SURF project include:
- Modeling and simulations of cell-free (sub)systems and synthetic cells (vesicles) consisting of cell-free extracts and circuits.
- Using some of the cell-free extract and TX-TL data, modeling and identifying model parameters using parameter identification tools. Studying structural parameter identifiability for these nonlinear models is another related direction.
- For a given circuit model, using tools such as global sensitivity analysis and parameter identifiability analysis to propose a decomposition of the circuit using the model so that methodical system identification by parts can be performed. A related direction of research could be to study reduced order models and their mapping back and forth to full order models.
We are interested in both theoretical and computational directions for this project. Experience with programming in Python and an understanding of feedback control systems are a bonus.
References:
- Hsiao, Victoria, Anandh Swaminathan, and Richard M. Murray. "Control theory for synthetic Biology: Recent advances in system characterization, control design, and controller implementation for synthetic biology." IEEE Control Systems Magazine 38.3 (2018): 32-62.
- Del Vecchio, Domitilla, Aaron J. Dy, and Yili Qian. "Control theory meets synthetic biology." Journal of The Royal Society Interface 13.120 (2016): 20160380.
- Tuza, Zoltan A., et al. "An in silico modeling toolbox for rapid prototyping of circuits in a biomolecular “breadboard” system." 52nd IEEE Conference on Decision and Control. IEEE, 2013. Link
- Myers, Chris J., et al. "iBioSim: a tool for the analysis and design of genetic circuits." Bioinformatics 25.21 (2009): 2848-2849.
- Swaminathan, Anandh, et al. "Fast and flexible simulation and parameter estimation for synthetic biology using bioscrape." (2017). Github link
- BioCRNPyler - Biomolecular Chemical Reaction Network Compiler : A Python toolbox to create CRN models in SBML for biomolecular mechanisms. Github link
- Sub-SBML : A Python based toolbox to create, edit, combine, and model interactions among multiple Systems Biology Markup Language (SBML) models. Github link