SURF 2021: Modeling tools for design and analysis of synthetic biological circuits
From Murray Wiki
Jump to navigationJump to search
SURF 2020 project description
- Mentor: Richard Murray
- Co-mentor: Ayush Pandey
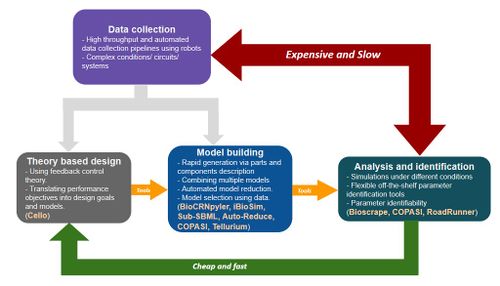
Intro about modeling in synthetic biology and the pipeline in the figure.
Intro about the project(s) possible.
Particular details on modeling work going on in the lab.
Research directions for the SURF project include:
- Modeling of cell-free systems
- System identification by parts, global sensitivity analysis, model decomposition, reduced models
- Using extract and tx-tl data
- Subsystem modeling of a build a cell project
We are interested in both theoretical and computational directions for this project. Experience with programming in Python and an understanding of feedback control systems are a bonus.
References:
- Hsiao, Victoria, Anandh Swaminathan, and Richard M. Murray. "Control theory for synthetic Biology: Recent advances in system characterization, control design, and controller implementation for synthetic biology." IEEE Control Systems Magazine 38.3 (2018): 32-62.
- Del Vecchio, Domitilla, Aaron J. Dy, and Yili Qian. "Control theory meets synthetic biology." Journal of The Royal Society Interface 13.120 (2016): 20160380.
- Tuza, Zoltan A., et al. "An in silico modeling toolbox for rapid prototyping of circuits in a biomolecular “breadboard” system." 52nd IEEE Conference on Decision and Control. IEEE, 2013. Link
- Swaminathan, Anandh, et al. "Fast and flexible simulation and parameter estimation for synthetic biology using bioscrape." (2017). Github link
- BioCRNPyler - Biomolecular Chemical Reaction Network Compiler : A Python toolbox to create CRN models in SBML for biomolecular mechanisms. Github link
- Sub-SBML : A Python based toolbox to create, edit, combine, and model interactions among multiple Systems Biology Markup Language (SBML) models. Github link