SURF 2021: Modeling tools for design and analysis of synthetic biological circuits
From Murray Wiki
Jump to navigationJump to search
SURF 2020 project description
- Mentor: Richard Murray
- Co-mentor: Ayush Pandey
Research directions for the SURF project include:
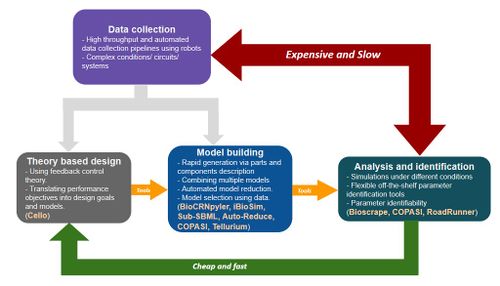
- Modeling of cell-free systems
- System identification by parts, global sensitivity analysis, model decomposition, reduced models
- Using extract and tx-tl data
We are interested in both theoretical and computational directions for this project.
References:
- Tuza, Zoltan A., et al. "An in silico modeling toolbox for rapid prototyping of circuits in a biomolecular “breadboard” system." 52nd IEEE Conference on Decision and Control. IEEE, 2013. Link
- Bioscrape
- BioCRNpyler
- Sub-SBML