SURF 2020: Applications of synthetic differentiation
2020 SURF project description
- Mentor: Richard Murray
- Co-mentor: Rory Williams
Applications of synthetic differentiation
In the Murray lab, we have demonstrated that the evolutionary stability of burdensome functions in E. coli can be improved by implementing tunable synthetic differentiation circuits utilizing serine integrases. Specifically, a population of progenitor cells which encodes, but does not express, the desired function, is able to replicate the DNA encoding the engineered function in the absence of its associated burden. An integrase-mediated genomic recombination event in a progenitor cell then both activates the engineered function by initiating the expression of T7RNAP, and starts a timer to limit the number of divisions differentiated cells are able to undergo. By appropriately tuning the differentiation rate through the inducible expression of degradation-tagged integrase protein, both the duration of circuit function and the total production achieved can be improved relative to the naïve implementation of the engineered function where all cells constitutively express the function. Though moderately effective, this strategy ultimately fails due to the selection of mutations which destroy the differentiation mechanism, and we are currently developing and testing circuits which will require two or more mutations to destroy the differentiation mechanism in order to improve its performance
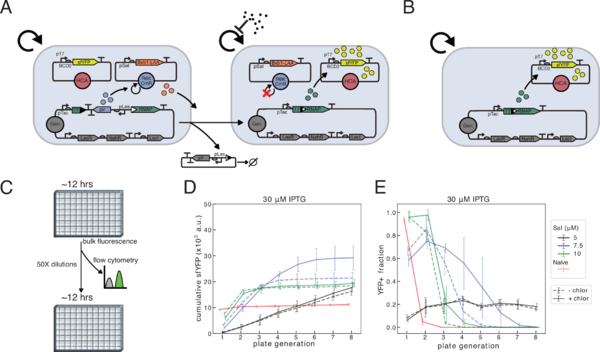
Figure 1: (A) Schematic of differentiation activated T7RNAP driven expression. Las AHL induces expression of pir protein, which is needed for replication of the R6K plasmid. Salicylate induces the expression of degradation-tagged Bxb1 integrase, which excises the pir expression cassette from the genome, and activates the expression of T7RNAP by bringing together two sections of the coding sequence. IPTG induces expression of full length T7RNAP in differentiated cells, allowing expression of sfYFP from the high-copy Cole1 plasmid. (B) Schematic of naïve inducible T7RNAP driven expression. Circuit is identical to a differentiated cell, but lacking the R6K plasmid. (C) Schematic for long-term differentiation experiments. (D-E) JS006 with differentiation circuit (A) above grown in M9CA + carbenicillin/3M Las AHL, +/- chloramphenicol, 30M IPTG, in varying concentrations of salicylate. Cells with naïve circuit (B) are grown in M9CA + carbenicillin/30M IPTG. Cells diluted 50X into same media conditions into 300L total volume every ~12 h for 8 total growths. (D) Cumulative total production plotted is the sum of endpoint fluorescence values from 12 h plate reader growth. (E) Samples taken immediately after growth were analyzed by flow cytometry, and fraction sfYFP positive cells is plotted. (D-E) Means of four total replicates from two independent experiments with standard deviation error bars.
We are interested in expanding the scope of differentiation architectures in a variety of ways, any of which could provide an opportunity for a SURF project.
1) Differentiation with reproductive and metabolic division of labor
While thus far we have been implementing differentiation in the context single-celled planktonically growing E. coli, naturally occurring examples of microbial differentiation are predominantly or exclusively found in species which form multicellular aggregates. In the context of multicellular aggregates, metabolic labor can be divided while allowing the benefits of this division of labor to be shared locally within the aggregate, and not with non-differentiating ‘cheaters’ or other competing microbes. We are currently exploring implementing differentiation in E. coli which form multicellular aggregates (through a combination of genetically controlled septation, and expression of pairs of cell-surface proteins facilitating cell-cell adhesion), and are interested in the potential of metabolic division of labor to increase the evolutionary stability of our differentiation architecture. This division of labor could take the form of amino acid cross-feeding, engineered nitrogen-fixation, etc.
2) Spatial patterning of multicellular E. coli
What types of spatial patterning can you achieve by combining synthetic differentiation with quorum-sensing/cell-cell communication, and the ability to genetically control cell lysis, septation and cell-cell adhesion? Can you make filamentous E. coli with regular patterns of distinct cell types? Can you genetically program the number of cells in these filamentous E. coli? Can you program the development of a hollow sphere of cells?
3) Production of burdensome or toxic proteins or metabolites using differentiation circuits
In the characterization of the capacity of differentiation circuits to improve the evolutionary stability of burdensome functions, we have exclusively used overproduction of a fluorescent protein. However, this circuit should be able to facilitate the production of any burdensome or even toxic protein or metabolite, particularly if the toxicity is specific to producer cells and not to all cells in the vessel. To explore this, a SURF student could identify useful/valuable protein(s) or metabolite(s) that currently are difficult to produce effectively in live cells, and evaluate their ability to be produced using differentiation circuits.
Useful previous experience
- molecular biology (pipetting, PCR, bacterial transformation, minipreps, etc.)
- coding (python preferred)
- mathematical modeling
Relevant publications
- Williams, R. L.; Murray, R. M. Tunable Integrase-Mediated Differentiation Facilitates Improved Output of Burdensome Functions in E. Coli. bioRxiv 2019. https://doi.org/10.1101/614529.
- Glass, D. S.; Riedel-Kruse, I. H. A Synthetic Bacterial Cell-Cell Adhesion Toolbox for Programming Multicellular Morphologies and Patterns. Cell 2018, 174 (3), 649-658.e16. https://doi.org/10.1016/j.cell.2018.06.041.
- Van Gestel, J.; Vlamakis, H.; Kolter, R. Division of Labor in Biofilms: The Ecology of Cell Differentiation. Microbiol. Spectr. 2015, 3 (2), 1–24. https://doi.org/10.1128/microbiolspec.mb-0002-2014.
- Wahl, M. E.; Murray, A. W. Multicellularity Makes Somatic Differentiation Evolutionarily Stable. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (30), 8362–8367. https://doi.org/10.1073/pnas.1608278113.