SURF 2019: Engineering a synthetic high-bandwidth intercellular communication system through plasmid conjugation
2019 SURF project description
- Mentor: Richard Murray
- Co-mentor: John Marken
Background
In order to realize many of synthetic biology's long-term goals, it is necessary to engineer populations of interacting cells rather than focusing on engineering individual cells alone. So far, work in this area has almost exclusively used quorum sensing systems to transfer information between cells, due to these systems' simplicity and modularity. However, because a quorum sensing system encodes its information in the concentration of a single signaling molecule, and because different systems exhibit a high degree of cross-talk with each other, quorum sensing is a low-bandwidth information transmission medium. This makes quorum sensing a poor choice for implementing engineered populations with a high degree of complexity.
Plasmid conjugation is a common type of bacterial horizontal gene transfer where a plasmid in one cell transfers a copy of itself into another adjacent cell. Because entire synthetic circuits can be contained on the transferred plasmid, conjugation is an extremely high-bandwidth channel for intercellular information transfer. Furthermore, synthetic conjugation systems have already been developed where 'helper plasmids' encode all of the genes required for plasmid transfer but lack the ability to transfer themselves, thereby conferring on its host cell the ability to modularly send any plasmid that contains the appropriate 'origin of transfer' (oriT) sequence. Such systems allow distinctions to be made between 'sender' and 'receiver' strains, which is an important prerequisite in the development of a flexible intercellular communication system.
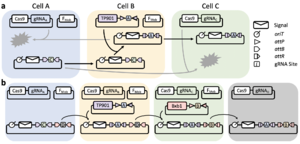
Despite the advantages of plasmid conjugation, so far there has been no published work which uses it as the basis for a general synthetic intercellular communication system. I am currently developing a 'message routing' framework that allows messages to be transferred only to designated recipients within a population. In this system, the cells express Cas9 and a guide RNA which serves to identify their strain. The signal plasmid contains an 'address' region which includes binding sites for the guide RNAs. The signal plasmid can only be transferred to a strain which does not have a site on the address, as otherwise the plasmid will be immediately cleaved by Cas9 upon receipt. Integrase attachment sites flank the guide RNA binding sites in the address, allowing the cells to use integrase-mediated cassette exchange to dynamically swap out guide RNA binding sites within the address, editing the list of 'allowed' recipients of the signal plasmid. In this way one can construct a defined path of information flow through a population, such as the linear path shown in Fig. 1. This message routing framework is just one example of the new capacities which are made available by conjugation-based signaling.
Project Goals
The aim of this project is to make experimental and/or theoretical contributions to the development of a conjugation-based intercellular communication system. These contributions can encompass anything from improving the fundamental properties of the conjugation system itself, to implementing a new population circuit that uses conjugation, to exploring broader concepts about what can be enabled by high-bandwidth intercellular communication. Some examples for project directions are given below.
Fruitful avenues for experimental work include:
- Creating helper plasmids with externally-controllable 'dials' to dynamically tune the rate of conjugation
- Implementing systems that allow cells to dynamically switch (and be switched) between sending and receiving states
- Creating spatially-defined population-level circuits
Interesting questions to address theoretically/computationally include:
- Are there new types of population-level circuits which can be built when plasmids (instead of quorum sensing signals) are the medium of communication?
- Can we think of general schemes for information representation in plasmids that allow for the encoding and interpretation of arbitrary messages? What are the advantages and disadvantages of different schemes?
Prerequisite Skills
For Experimental Work
- Familiarity with molecular cloning techniques and standard
BSL-1 wetlab procedures
- Familiarity with synthetic biology and genetic circuit design
For Computational Work
- Coding experience, preferably in Python
- Familiarity with differential equations
- Familiarity with synthetic biology and genetic circuit design
References