SURF 2015: Designing biomolecular temperature sensors
2015 SURF project description
- Mentor: Richard Murray
- Co-mentor: Shaunak Sen
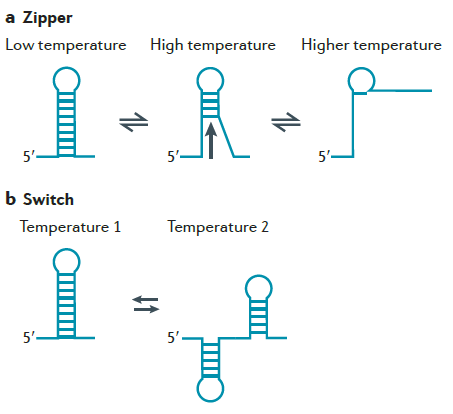
Certain types of biomolecular circuits have naturally evolved to respond to changes in temperature. For biomolecular circuit design, it is desired to develop toolbox of temperature sensors that have different levels of sensitivities and responses to temperature changes.
To address this, we plan to focus on specific RNA sequences, also called RNA thermometers, which, when placed upstream of a gene, have been demonstrated to increase gene expression as temperature increases. In preliminary work, we have generated a library of RNA thermometers that can generate diverse responses to temperature.
Based on this library, the goals of the project are twofold, 1. To quantitatively characterize this temperature dependence experimentally. This will include using measurements in cells and cell extracts (TX-TL) as well as exploring other methods to characterize temperature dependence. 2. Use these RNA thermometers to trigger other biomolecular circuits in a temperature-dependent fashion. For example, using RNA thermometer as an input to a pulse generating Feedforward Loop circuit can result in a temperature-dependent pulse.
Applications of this work should include efficient control of large volume chemical reactors, precise spatiotemporal control of gene expression as well as tools to design robustness to temperature in biomolecular circuits.