Physical Biology Boot Camp
These are notes from the Physical Biology Boot Camp that I participated in on 8-14 Oct 06 at Caltech.
About the boot camp
Participants
Instructors
- Rob Phillips (Caltech ME/APh)
- Hernan Garcia (Caltech Ph)
- Tristan Ursell (Caltech APh)
- Paul Grayson
- Lin Han
- Heun Jin Lee
- Eric Peterson
- Frosso Seitaridou
- Dave Wu
- Tabita Winther
Recruits
|
Visiting faculty
Others
|
Organization
There are basically four parts of boot camp:
- DNA Science - learning how to manipulate DNA
- Size of things - learning the various spatial scales in biology
- Rate of things - learning the various temporal scales in biology
- Project - a small project, working with a graduate student or visitor
Size of things
In the "size of things" section of the boot camp, we learned how to use microscopes to take (calibrated) measurements of various cells.
Rate of things
DNA Science
The DNA science section of bootcamp involved learning how to do cloning in E. coli. The basic idea of the project that we undertook was to place a fluorescent gene into a plasmid. We started with the pZS25 plasmid, which is a modular plasmid. Here is now to read the "part number" (Lutz and Bujard, 1997):
| pZ | Origin of replication | resistance marker | regulatory unit | |||
| E | ColE1 | 1 | Ampicilin | 1 | P_LtetO1-1 | |
| A | p15A | 2 | Kanemycin | 2 | P_LlacO1-1 | |
| S | pSC101 | 3 | Chloramphenicol | 3 | P_AllacO1-1 | |
| S* | pSC101* | 4 | Spectinomycin | 1 | P_lac/ara-1 | |
| 1 | Ampicilin | 5 | P_lacUV5-1 | |||
So pZS25 uses an SC101 origin of replication (low copy number; about 10 copies), has kanemycin antibiotic resistance, and uses P_lacUV5-1 as its regulatory unit (repressed by LacI and induced by IPTG).
The actual plasmid that we used was a version that had been modified by Hernan Garcia so that the promotor region had modifications that could be used to study the structural properties of DNA. Hernan created plasmids of the form pZS25.TA-94-YFP, which is a plasmid with a promotor region of length 94 (default), using a certain combination of sequences between the operator regions for lacI. The cloning that we want to do is to replace YFP with lacZ (which we extracted from a pZE21 plasmid).
Step 1: create pZS25 vector (Sun afternoon) - the first thing that we did was to use restriction enzymes to create our "vector", where we would eventually insert the lacZ gene. We did this but cutting the pZS25 plasmid using HindIII and KpmI in a "double digest". To get some sense of how the restriction enzymes worked, we did two controls as well: a KpmI contro and a HindIII control. In all, we generated four mixtures:
- Double digest
- KpmI control
- HindIII control
- Uncut plasmid
Step 2: extract lacZ from pZE21 (Sun afternoon) - to remove lacZ from the pZE21 plasmid, we used the HindIII and EcoR1 restriction enzymes. Hernan had already done the HindIII digest, so we only had to do ecoR1. For reasons that don't make complete sense to me, this DNA fragment was called lambda + HindIII.
Step 3: run gel and extract DNA (Sun afternoon) - to extract out the DNA that we created, we ran a gel. Here's a picture of Richard and Albert's gel:
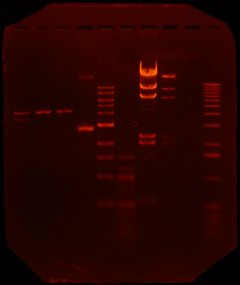
The first four tracks correspond to the DNA strands listed above. The fifth and sixth tracks are 1k bp and 100 bp ladders. Tracks 7 and 8 correspond to lambda + HindIII and lambda + HindIII + EcoR1. The 9th and 10th tracks were test tracks (just dye, I think).
Step 3: PCR amplification (Mon morning) - the next thing we did was to